- 著者
- Tomoki Nakayoshi Koichi Kato Eiji Kurimoto Akifumi Oda
- 出版者
- The Pharmaceutical Society of Japan
- 雑誌
- Biological and Pharmaceutical Bulletin (ISSN:09186158)
- 巻号頁・発行日
- vol.44, no.7, pp.967-975, 2021-07-01 (Released:2021-07-01)
- 参考文献数
- 39
- 被引用文献数
- 1
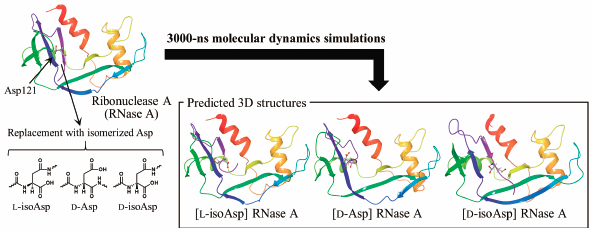
Isomerized aspartic acid (Asp) residues have previously been identified in various aging tissues, and are suspected to contribute to age-related diseases. Asp-residue isomerization occurs nonenzymatically under physiological conditions, resulting in the formation of three types of isomerized Asp (i.e., L-isoAsp, D-Asp, and D-isoAsp) residues. Asp-residue isomerization often accelerates protein aggregation and insolubilization, making structural biology analyses difficult. Recently, Sakaue et al. reported the synthesis of a ribonuclease A (RNase A) in which Asp121 was artificially replaced with different isomerized Asp residues, and experimentally demonstrated that the enzymatic activities of these artificial mutants were completely lost. However, their structural features have not yet been elucidated. In the present study, the three-dimensional (3D) structures of these artificial-mutant RNases A were predicted using molecular dynamics (MD) simulations. The 3D structures of wild-type and artificial-mutant RNases A were converged by 3000-ns MD simulations. Our computational data show that the structures of the active site and the formation frequencies of the appropriate catalytic dyad structures in the artificial-mutant RNases A were quite different from wild-type RNase A. These computational findings may provide an explanation for the experimental data which show that artificial-mutant RNases A lack enzymatic activity. Herein, MD simulations have been used to evaluate the influences of isomerized Asp residues on the 3D structures of proteins.