- 著者
- Kazunori Miwa Yan Guo Masayuki Hata Yoshinori Hirano Norio Yamamoto Tyuji Hoshino
- 出版者
- The Pharmaceutical Society of Japan
- 雑誌
- Chemical and Pharmaceutical Bulletin (ISSN:00092363)
- 巻号頁・発行日
- vol.71, no.12, pp.897-905, 2023-12-01 (Released:2023-12-01)
- 参考文献数
- 40
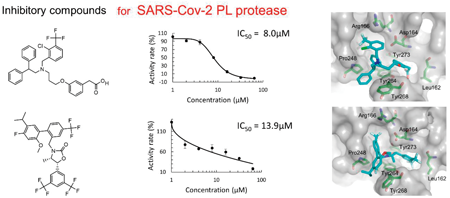
Virtual screening with high-performance computers is a powerful and cost-effective technique in drug discovery. A chemical database is searched to find candidate compounds firmly bound to a target protein, judging from the binding poses and/or binding scores. The severe acute respiratory syndrome coronavirus 2 (SARS-Cov-2) infectious disease has spread worldwide for the last three years, causing severe slumps in economic and social activities. SARS-Cov-2 has two viral proteases: 3-chymotrypsin-like (3CL) and papain-like (PL) protease. While approved drugs have already been released for the 3CL protease, no approved agent is available for PL protease. In this work, we carried out in silico screening for the PL protease inhibitors, combining docking simulation and molecular mechanics calculation. Docking simulations were applied to 8,820 molecules in a chemical database of approved and investigational compounds. Based on the binding poses generated by the docking simulations, molecular mechanics calculations were performed to optimize the binding structures and to obtain the binding scores. Based on the binding scores, 57 compounds were selected for in vitro assay of the inhibitory activity. Five inhibitory compounds were identified from the in vitro measurement. The predicted binding structures of the identified five compounds were examined, and the significant interaction between the individual compound and the protease catalytic site was clarified. This work demonstrates that computational virtual screening by combining docking simulation with molecular mechanics calculation is effective for searching candidate compounds in drug discovery.
- 著者
- Kazunori Miwa Yan Guo Masayuki Hata Norio Yamamoto Tyuji Hoshino
- 出版者
- The Pharmaceutical Society of Japan
- 雑誌
- Chemical and Pharmaceutical Bulletin (ISSN:00092363)
- 巻号頁・発行日
- vol.71, no.5, pp.360-367, 2023-05-01 (Released:2023-05-01)
- 参考文献数
- 39
- 被引用文献数
- 4
Computational screening is one of the fundamental techniques in drug discovery. Each compound in a chemical database is bound to the target protein in virtual, and candidate compounds are selected from the binding scores. In this work, we carried out combinational computation of docking simulation to generate binding poses and molecular mechanics calculation to estimate binding scores. The coronavirus infectious disease has spread worldwide, and effective chemotherapy is strongly required. The viral 3-chymotrypsin-like (3CL) protease is a good target of low molecular-weight inhibitors. Hence, computational screening was performed to search for inhibitory compounds acting on the 3CL protease. As a preliminary assessment of the performance of this approach, we used 51 compounds for which inhibitory activity had already been confirmed. Docking simulations and molecular mechanics calculations were performed to evaluate binding scores. The preliminary evaluation suggested that our approach successfully selected the inhibitory compounds identified by the experiments. The same approach was applied to 8820 compounds in a database consisting of approved and investigational chemicals. Hence, docking simulations, molecular mechanics calculations, and re-evaluation of binding scores including solvation effects were performed, and the top 200 poses were selected as candidates for experimental assays. Consequently, 25 compounds were chosen for in vitro measurement of the enzymatic inhibitory activity. From the enzymatic assay, 5 compounds were identified to have inhibitory activities against the 3CL protease. The present work demonstrated the feasibility of a combination of docking simulation and molecular mechanics calculation for practical use in computational virtual screening.
- 著者
- Taichi Kamo Keiichi Kuroda Shota Kondo Usaki Hayashi Satoshi Fudo Tomoki Yoneda Akiko Takaya Michiyoshi Nukaga Tyuji Hoshino
- 出版者
- The Pharmaceutical Society of Japan
- 雑誌
- Chemical and Pharmaceutical Bulletin (ISSN:00092363)
- 巻号頁・発行日
- vol.69, no.12, pp.1179-1183, 2021-12-01 (Released:2021-12-01)
- 参考文献数
- 29
- 被引用文献数
- 3
Metallo-β-lactamases (MBLs) are significant threats to humans because they deteriorate many kinds of β-lactam antibiotics and are key enzymes responsible for multi-drug resistance of bacterial pathogens. As a result of in vitro screening, two compounds were identified as potent inhibitors of two kinds of MBLs: imipenemase (IMP-1) and New Delhi metallo-β-lactamase (NDM-1). The binding structure of one of the identified compounds was clarified by an X-ray crystal analysis in complex with IMP-1, in which two possible binding poses were observed. Molecular dynamics (MD) simulations were performed by building two calculation models from the respective binding poses. The compound was stably bound to the catalytic site during the simulation in one pose. The binding model between NDM-1 and the compound was constructed for MD simulation. Calculation results for NDM-1 were similar to those of IMP-1. The simulation suggested that the binding of the identified inhibitory compound was also durable in the catalytic site of NDM-1. The compound will be a sound basis for the development of the inhibitors for MBLs.