- 著者
- Akihiro Ito Lei Wang Ryotaro Notomi Shigeki Sasaki Yosuke Taniguchi
- 出版者
- The Pharmaceutical Society of Japan
- 雑誌
- Chemical and Pharmaceutical Bulletin (ISSN:00092363)
- 巻号頁・発行日
- vol.72, no.1, pp.16-20, 2024-01-01 (Released:2024-01-01)
- 参考文献数
- 24
Triplex DNA formation has generated much interest as a genomic targeting tool that directly targets duplex DNA. However, fundamental limitations in the base pairs of target duplex DNA sequences that can form stable triplex DNA have limited the application. Recently, we have reported on the recognition of CG and 5mCG base pairs by artificial nucleic acid derivatives with a 2′-deoxynebularine skeleton. Therefore, we attempted to explore the basic skeleton that is important for the development of new artificial nucleic acids allowing for the recognition of TA base pairs. In this study, we focused on a benzimidazole skeleton and introduced a hydroxyl group to enable one-point hydrogen bonding. We have synthesized artificial nucleoside analogues with hydroxyl group on the benzimidazole and incorporated their amidite derivatives into triplex forming oligonucleotides (TFOs). The gel shift assay was performed to evaluate the triplex DNA formation ability of synthesized TFOs, and TFOs containing hydroxybenzimidazole were successfully recognized TA base pairs for all four different sequences. Moreover, compared to the results for the TFOs containing benzimidazole, which suggested hydrogen bonding formation at the hydroxyl group. Therefore, hydroxybenzimidazole would be an important artificial nucleic acid skeleton for TA base pair recognition.
1 0 0 0 OA Site-Specific Tritium Labeling at the Predefined Internal Position of the Chemically-Modified RNA
- 著者
- Hirotaka Murase Jeongsu Lee Yosuke Taniguchi Shigeki Sasaki
- 出版者
- The Pharmaceutical Society of Japan
- 雑誌
- Chemical and Pharmaceutical Bulletin (ISSN:00092363)
- 巻号頁・発行日
- vol.71, no.1, pp.64-69, 2023-01-01 (Released:2023-01-01)
- 参考文献数
- 16
- 被引用文献数
- 1
In nucleic acid drug discovery, it is extremely important to develop a technology to understand the distribution in target organs and to trace the degradation process in the body in order to optimize the structure and improve the efficiency of the clinical trial process. Since nucleic acid drugs are essentially metabolically degraded into numerous fragments, labeling at the internal position is preferable to that at the terminus. Due to the high molar specific activity of tritium, various approaches for tritium-labeling have been studied for nucleic acid drugs. Nevertheless, a generally-applicable method for tritium labeling of the internal position of a nucleic acid has not been established. In this study, we have demonstrated a new and efficient method for site-specific tritium labeling of the cytosine base at a predefined internal position in nucleic acid drugs. This method was developed by the chemical modification of the cytosine 4-amino group with the pyridinyl vinyl keto group by the functionality-transfer reaction using the reactive oligodeoxynucleotide (ODN), followed by reduction with NaBT4. Applicability to a variety of chemical structures, such as 5-methyl cytosine, 2′-O-methyl, 2′-fluoro ribose derivatives, Locked/Bridged nucleic acid (LNA/BNA) derivatives, as well as phosphorothioate bonds, has been evidenced using nine oligoribonucleic acid (ORN) substrates. It has been clearly demonstrated that this method is an excellent method for tritium-labeling of nucleic acid with an average conversion efficiency of 74%, an average isolated labeling yield of 60%, and an average specific activity of 61 GBq/mmol. This method is expected to contribute to the preclinical absorption, distribution, metabolism, excretion (ADME) studies of nucleic acid drug candidates.
- 著者
- Norihiro Togo Hirotaka Murase Jeongsu Lee Yosuke Taniguchi Shigeki Sasaki
- 出版者
- The Pharmaceutical Society of Japan
- 雑誌
- Chemical and Pharmaceutical Bulletin (ISSN:00092363)
- 巻号頁・発行日
- vol.70, no.7, pp.498-504, 2022-07-01 (Released:2022-07-01)
- 参考文献数
- 17
- 被引用文献数
- 2
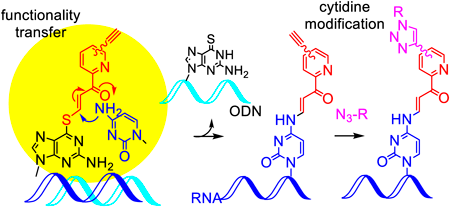
Due to the importance of the RNA chemical modifications, methods for the selective chemical modification at a predetermined site of the internal position of RNA have attracted much attention. We have developed functional artificial nucleic acids that modify a specific site of RNA in a site- and base-selective manner. In addition, the copper-catalyzed azide-alkyne cycloaddition (CuAAC) has been shown to introduce additional molecules on the alkynes attached to the pyridine ring. However, it was found that some azide compounds produced the cycloadduct in lower yields. Therefore, in this study, we synthesized the pyridinyl transfer group with the alkyne attached via a polyethylene glycol (PEG) linker with a different length and optimized its structure for both the transfer and CuAAC reaction. Three new transfer groups were synthesized by introducing an alkyne group at the end of the triethylene (11), tetraethylene (12) or pentaethylen glycol linker (13) at the 5-position of the pyridine ring of (E)-3-iodo-1-(pyridin-2-yl)prop-2-en-1-one. These transfer groups were introduced to the 6-thioguanine base in the oligodeoxynucleotide (ODN) in high yields. The transfer groups 11 and 12 more efficiently underwent the cytosine modification. For the CuAAC reaction, although 7 showed low adduct yields with the anionic azide compound, the new transfer groups, especially 12 and 13, significantly improved the yields. In conclusion, the transfer groups 12 and 13 were determined to be promising compounds for the modification of long RNAs.