- 著者
- Keisuke Arikawa
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.15, pp.58-74, 2018 (Released:2018-02-27)
- 参考文献数
- 50
- 被引用文献数
- 2
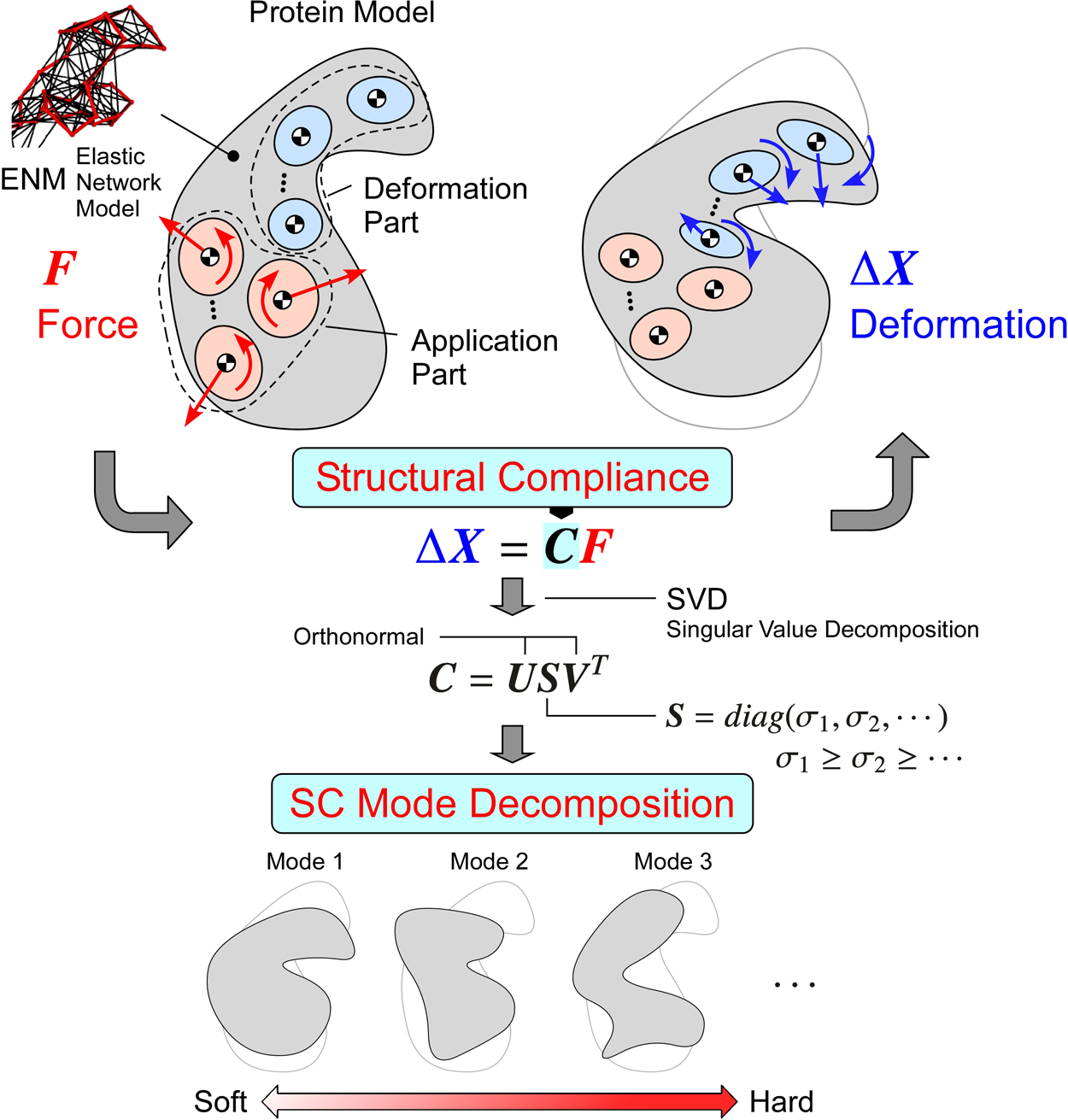
We propose methods for directly analyzing structural compliance (SC) properties of elastic network models of proteins, and we also propose methods for extracting information about motion properties from the SC properties. The analysis of SC properties involves describing the relationships between the applied forces and the deformations. When decomposing the motion according to the magnitude of SC (SC mode decomposition), we can obtain information about the motion properties under the assumption that the lower SC mode motions or the softer motions occur easily. For practical applications, the methods are formulated in a general form. The parts where forces are applied and those where deformations are evaluated are separated from each other for enabling the analyses of allosteric interactions between the specified parts. The parts are specified not only by the points but also by the groups of points (the groups are treated as flexible bodies). In addition, we propose methods for quantitatively evaluating the properties based on the screw theory and the considerations of the algebraic structures of the basic equations expressing the SC properties. These methods enable quantitative discussions about the relationships between the SC mode motions and the motions estimated from two different conformations; they also help identify the key parts that play important roles for the motions by comparing the SC properties with those of partially constrained models. As application examples, lactoferrin and ATCase are analyzed. The results show that we can understand their motion properties through their lower SC mode motions or the softer motions.