- 著者
- Takao K. Suzuki
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.19, pp.e190011, 2022 (Released:2022-04-20)
- 参考文献数
- 98
- 被引用文献数
- 3
Design principles of phenotypes in organisms are fundamental issues in physical biology. So far, understanding “systems” of living organisms have been chiefly promoted by understanding the underlying biomolecules such as genes and proteins, and their intra- and inter-relationships and regulations. After a long period of sophistication, biophysics and molecular biology have established a general framework for understanding ‘molecular systems’ in organisms without regard to species, so that the findings of fly studies can be applied to mouse studies. However, little attention has been paid to exploring “phenotypic systems” in organisms, and thus its general framework remains poorly understood. Here I review concepts, methods, and case studies using butterfly and moth wing patterns to explore phenotypes as systems. First, I present a unifying framework for phenotypic traits as systems, termed multi-component systems. Second, I describe how to define components of phenotypic systems, and also show how to quantify interactions among phenotypic parts. Subsequently, I introduce the concept of the macro-evolutionary process, which illustrates how to generate complex traits. In this point, I also introduce mathematical methods, “phylogenetic comparative methods”, which provide stochastic processes along molecular phylogeny as bifurcated paths to quantify trait evolution. Finally, I would like to propose two key concepts, macro-evolutionary pathways and genotype-phenotype loop (GP loop), which must be needed for the next directions. I hope these efforts on phenotypic biology will become one major target in biophysics and create the next generations of textbooks. This review article is an extended version of the Japanese article, Biological Physics in Phenotypic Systems of Living Organisms, published in SEIBUTSU-BUTSURI Vol. 61, p. 31–35 (2021).
30 0 0 0 OA Limitations of the ABEGO-based backbone design: ambiguity between αα-corner and αα-hairpin
- 著者
- Koya Sakuma
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.18, pp.159-167, 2021 (Released:2021-06-18)
- 参考文献数
- 23
ABEGO is a coarse-grained representation for polypeptide backbone dihedral angles. The Ramachandran map is divided into four segments denoted as A, B, E, and G to represent the local conformation of polypeptide chains in the character strings. Although the ABEGO representation is widely used in backbone building simulation for de novo protein design, it cannot capture minor differences in backbone dihedral angles, which potentially leads to ambiguity between two structurally distinct fragments. Here, I show a nontrivial example of two local motifs that could not be distinguished by their ABEGO representations. I found that two well-known local motifs αα-hairpins and αα-corners are both represented as α-GBB-α and thus indistinguishable in the ABEGO representation, although they show distinct arrangements of the flanking α-helices. I also found that α-GBB-α motifs caused a loss of efficiency in the ABEGO-based fragment-assembly simulations for de novo protein backbone design. Nevertheless, I was able to design amino-acid sequences that were predicted to fold into the target topologies that contained these α-GBB-α motifs, which suggests such topologies that are difficult to build by ABEGO-based simulations are designable once the backbone structures are modeled by some means. The finding that certain local motifs bottleneck the ABEGO-based fragment-assembly simulations for construction of backbone structures suggests that finer representations of backbone torsion angles are required for efficiently generating diverse topologies containing such indistinguishable local motifs.
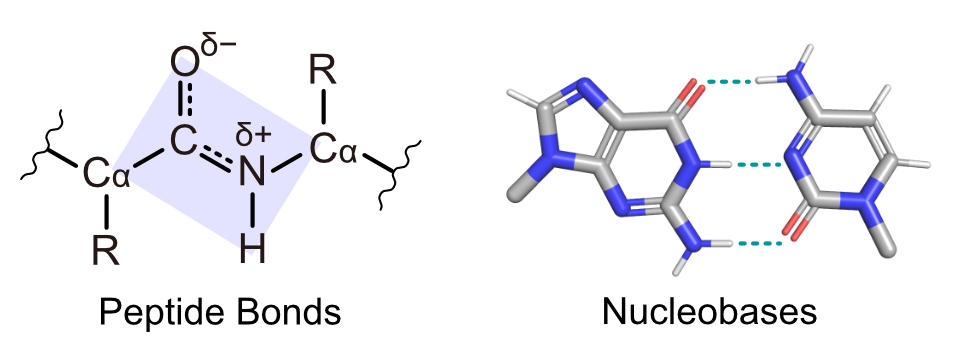
21 0 0 0 OA Why we are made of proteins and nucleic acids: Structural biology views on extraterrestrial life
- 著者
- Shunsuke Tagami
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.20, no.2, pp.e200026, 2023 (Released:2023-06-20)
- 参考文献数
- 109
Is it a miracle that life exists on the Earth, or is it a common phenomenon in the universe? If extraterrestrial organisms exist, what are they like? To answer these questions, we must understand what kinds of molecules could evolve into life, or in other words, what properties are generally required to perform biological functions and store genetic information. This review summarizes recent findings on simple ancestral proteins, outlines the basic knowledge in textbooks, and discusses the generally required properties for biological molecules from structural biology viewpoints (e.g., restriction of shapes, and types of intra- and intermolecular interactions), leading to the conclusion that proteins and nucleic acids are at least one of the simplest (and perhaps very common) forms of catalytic and genetic biopolymers in the universe. This review article is an extended version of the Japanese article, On the Origin of Life: Coevolution between RNA and Peptide, published in SEIBUTSU BUTSURI Vol. 61, p. 232–235 (2021).
- 著者
- Masami Ikeda Minoru Sugihara Makiko Suwa
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.15, pp.104-110, 2018 (Released:2018-04-27)
- 参考文献数
- 28
- 被引用文献数
- 5
We report the development of the SEVENS database, which contains information on G-protein coupled receptor (GPCR) genes that are identified with high confidence levels (A, B, C, and D) from various eukaryotic genomes, by using a pipeline comprising bioinformatics softwares, including a gene finder, a sequence alignment tool, a motif and domain assignment tool, and a transmembrane helix predictor.SEVENS compiles detailed information on GPCR genes, such as chromosomal mapping position, phylogenetic tree, sequence similarity to known genes, and protein function described by motif/domain and transmembrane helices. They are presented in a user-friendly interface. Because of the comprehensive gene findings from genomes, SEVENS contains a larger data set than that of previous databases and enables the performance of a genome-scale overview of all the GPCR genes. We surveyed the complete genomes of 68 eukaryotes, and found that there were between 6 and 3,470 GPCR genes for each genome (Level A data). Within these genes, the number of receptors for various molecules, including biological amines, peptides, and lipids, were conserved in mammals, birds, and fishes, whereas the numbers of odorant receptors and pheromone receptors were highly diverse in mammals. SEVENS is freely available at http://sevens.cbrc.jp or http://sevens.chem.aoyama.ac.jp.
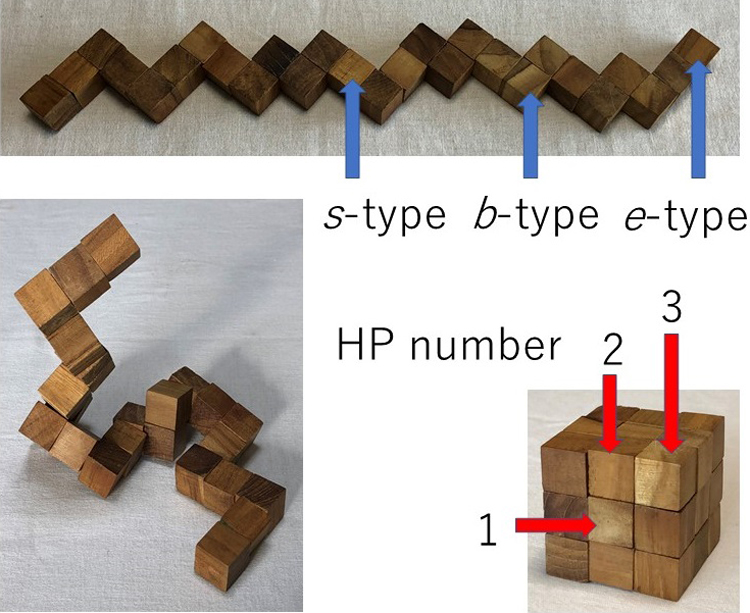
14 0 0 0 OA Snake cube puzzle and protein folding
- 著者
- Nobuhiro Go
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.16, pp.256-263, 2019 (Released:2019-11-29)
- 参考文献数
- 11
- 被引用文献数
- 5
The snake cube puzzle made of a linear array of 27 cubes and its modified and extended versions are used as theoretical models to study the mechanism of folding of proteins into their sequence-specific native three-dimensional structures. Each of the three versions is characterized by the respective set of characteristics attributed to each of its constituent cubes and an array is characterized by its specific sequence of the cube characteristics. The aim of the puzzles is to fold the cube array into a compact 3×3×3 cubic structure. In all three versions, out of all possible sequences, only a limited fraction of sequences are found foldable into the compact cube. Even among foldable sequences, the structures folded into the compact 3×3×3 cube are found often not uniquely determined from the sequence. By comparing the results obtained for the three versions of models, we conclude that the power of the hydrophobic interactions to make the folded structure unique to the sequence is much weaker than the geometrical varieties of constituent cubes as modelled in the original snake cube puzzle. However, when this weak cube attribute is compounded to that of the original snake cube puzzle, the power is enhanced very effectively. This is a strong manifestation of the consistency principle: The sequence-specific native structure of protein is realized as a result of consistency of various types of interactions working in protein.
- 著者
- Tetsuichi Wazawa Takeharu Nagai
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.e200030, (Released:2023-06-28)
Ion currents associated with channel proteins in the presence of membrane potential are ubiquitous in cellular and organelle membranes. When an ion current occurs through a channel protein, Joule heating should occur. However, this Joule heating seems to have been largely overlooked in biology. Here we show theoretical investigation of Joule heating involving channel proteins in biological processes. We used electrochemical potential to derive the Joule’s law for an ion current through an ion transport protein in the presence of membrane potential, and we suggest that heat production and absorption can occur. Simulation of temperature distribution around a single channel protein with the Joule heating revealed that the temperature increase was as small as <10−3 K, although an ensemble of channel proteins was suggested to exhibit a noticeable temperature increase. Thereby, we theoretically investigated the Joule heating of systems containing ensembles of channel proteins. Nerve is known to undergo rapid heat production followed by heat absorption during the action potential, and our simulation of Joule heating for a squid giant axon combined with the Hodgkin-Huxley model successfully reproduced the feature of the heat. Furthermore, we extended the theory of Joule heating to uncoupling protein 1 (UCP1), a solute carrier family transporter, which is important to the non-shivering thermogenesis in brown adipose tissue mitochondria (BATM). Our calculations showed that the Joule heat involving UCP1 was comparable to the literature calorimetry data of BATM. Joule heating of ion transport proteins is likely to be one of important mechanisms of cellular thermogenesis.
13 0 0 0 OA Why we are made of proteins and nucleic acids: Structural biology views on extraterrestrial life
- 著者
- Shunsuke Tagami
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.e200026, (Released:2023-06-02)
Is it a miracle that life exists on the Earth, or is it a common phenomenon in the universe? If extraterrestrial organisms exist, what are they like? To answer these questions, we must understand what kinds of molecules could evolve into life, or in other words, what properties are generally required to perform biological functions and store genetic information. This review summarizes recent findings on simple ancestral proteins, outlines the basic knowledge in textbooks, and discusses the generally required properties for biological molecules from structural biology viewpoints (e.g., restriction of shapes, and types of intra- and intermolecular interactions), leading to the conclusion that proteins and nucleic acids are at least one of the simplest (and perhaps very common) forms of catalytic and genetic biopolymers in the universe. This review article is an extended version of the Japanese article, On the Origin of Life: Coevolution between RNA and Peptide, published in SEIBUTSU BUTSURI Vol. 61, p. 232-235 (2021).
- 著者
- Hiroshi Imamura
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.e200021, (Released:2023-05-10)
- 被引用文献数
- 1
Small-angle scattering (SAS) is a powerful tool for the detailed structural analysis of objects at the nanometer scale. In contrast to techniques such as electron microscopy, SAS data are presented as reciprocal space information, which hinders the intuitive interpretation of SAS data. This study presents a workflow: (1) creating objects, (2) 3D scanning, (3) the representation of the object as point clouds on a laptop, (4) computation of a distance distribution function, and (5) computation of SAS, executed via the computer program Phone2SAS. This enables us to realize SAS and perform the interactive modeling of SAS of the object of interest. Because 3D scanning is easily accessible through smartphones, this workflow driven by Phone2SAS contributes to the widespread use of SAS. The application of Phone2SAS for the structural assignment of SAS to Y-shaped antibodies is reported in this study.
- 著者
- Taro Furubayashi Norikazu Ichihashi
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.e190005, (Released:2022-02-15)
- 著者
- Takeru Kameda Akinori Awazu Yuichi Togashi
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.19, pp.e190027, 2022 (Released:2022-09-08)
- 参考文献数
- 211
- 被引用文献数
- 2
With the recent progress in structural biology and genome biology, structural dynamics of molecular systems that include nucleic acids has attracted attention in the context of gene regulation. The structure–function relationship is an important topic that highlights the importance of the physicochemical properties of nucleotides, as well as that of amino acids in proteins. Simulations are a useful tool for the detailed analysis of molecular dynamics that complement experiments in molecular biology; however, molecular simulation of nucleic acids is less well developed than that of proteins partly due to the physical nature of nucleic acids. In this review, we briefly describe the current status and future directions of the field as a guide to promote collaboration between experimentalists and computational biologists.
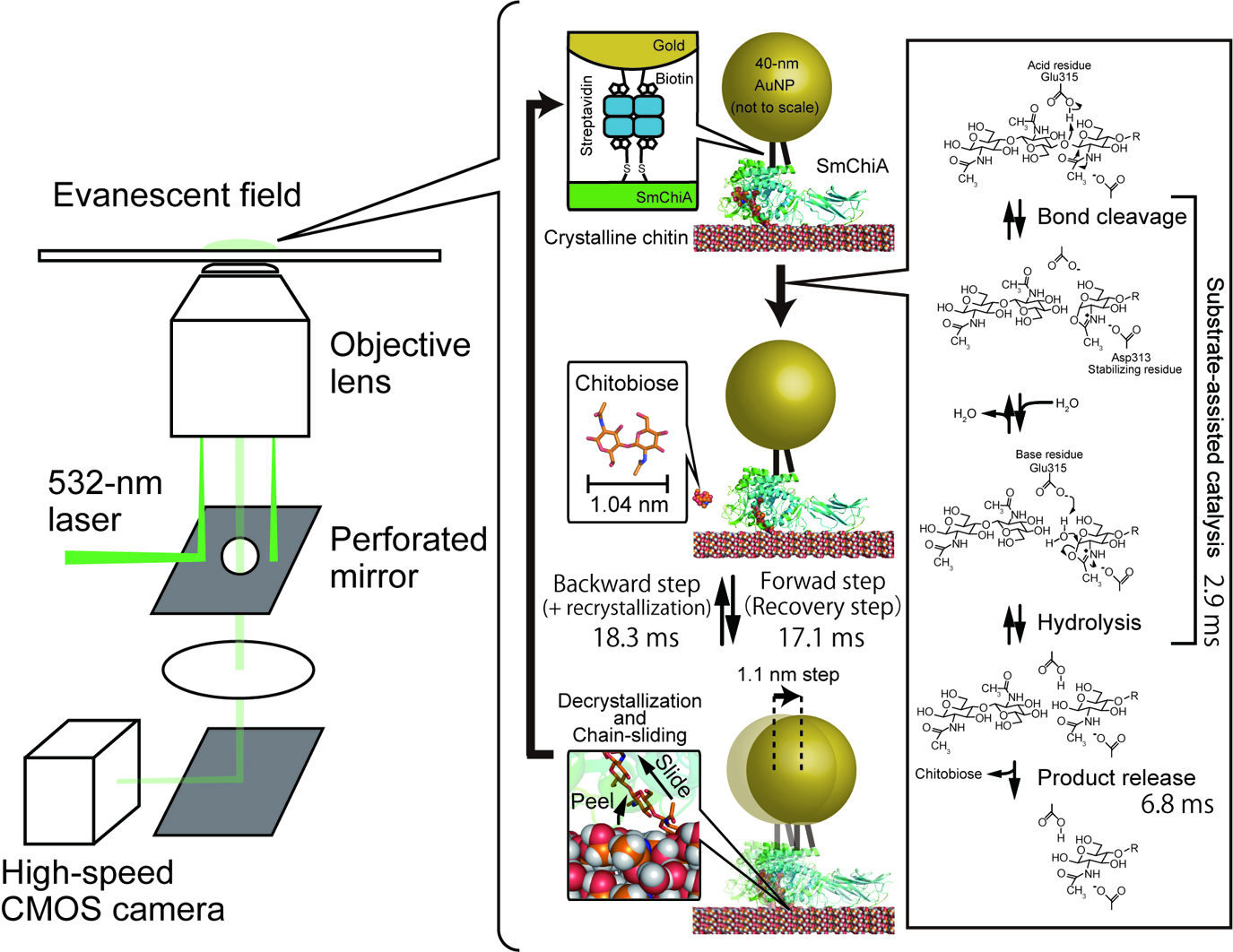
- 著者
- Akihiko Nakamura Kei-ichi Okazaki Tadaomi Furuta Minoru Sakurai Jun Ando Ryota Iino
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.17, pp.51-58, 2020 (Released:2020-07-10)
- 参考文献数
- 26
- 被引用文献数
- 4 5
Motor proteins are essential units of life and are well-designed nanomachines working under thermal fluctuations. These proteins control moving direction by consuming chemical energy or by dissipating electrochemical potentials. Chitinase A from bacterium Serratia marcescens (SmChiA) processively moves along crystalline chitin by hydrolysis of a single polymer chain to soluble chitobiose. Recently, we directly observed the stepping motions of SmChiA labeled with a gold nanoparticle by dark-field scattering imaging to investigate the moving mechanism. Time constants analysis revealed that SmChiA moves back and forth along the chain freely, because forward and backward states have a similar free energy level. The similar probabilities of forward-step events (83.5%=69.3%+14.2%) from distributions of step sizes and chain-hydrolysis (86.3%=(1/2.9)/(1/2.9+1/18.3)×100) calculated from the ratios of time constants of hydrolysis and the backward step indicated that SmChiA moves forward as a result of shortening of the chain by a chitobiose unit, which stabilizes the backward state. Furthermore, X-ray crystal structures of sliding intermediate and molecular dynamics simulations showed that SmChiA slides forward and backward under thermal fluctuation without large conformational changes of the protein. Our results demonstrate that SmChiA is a burnt-bridge Brownian ratchet motor.
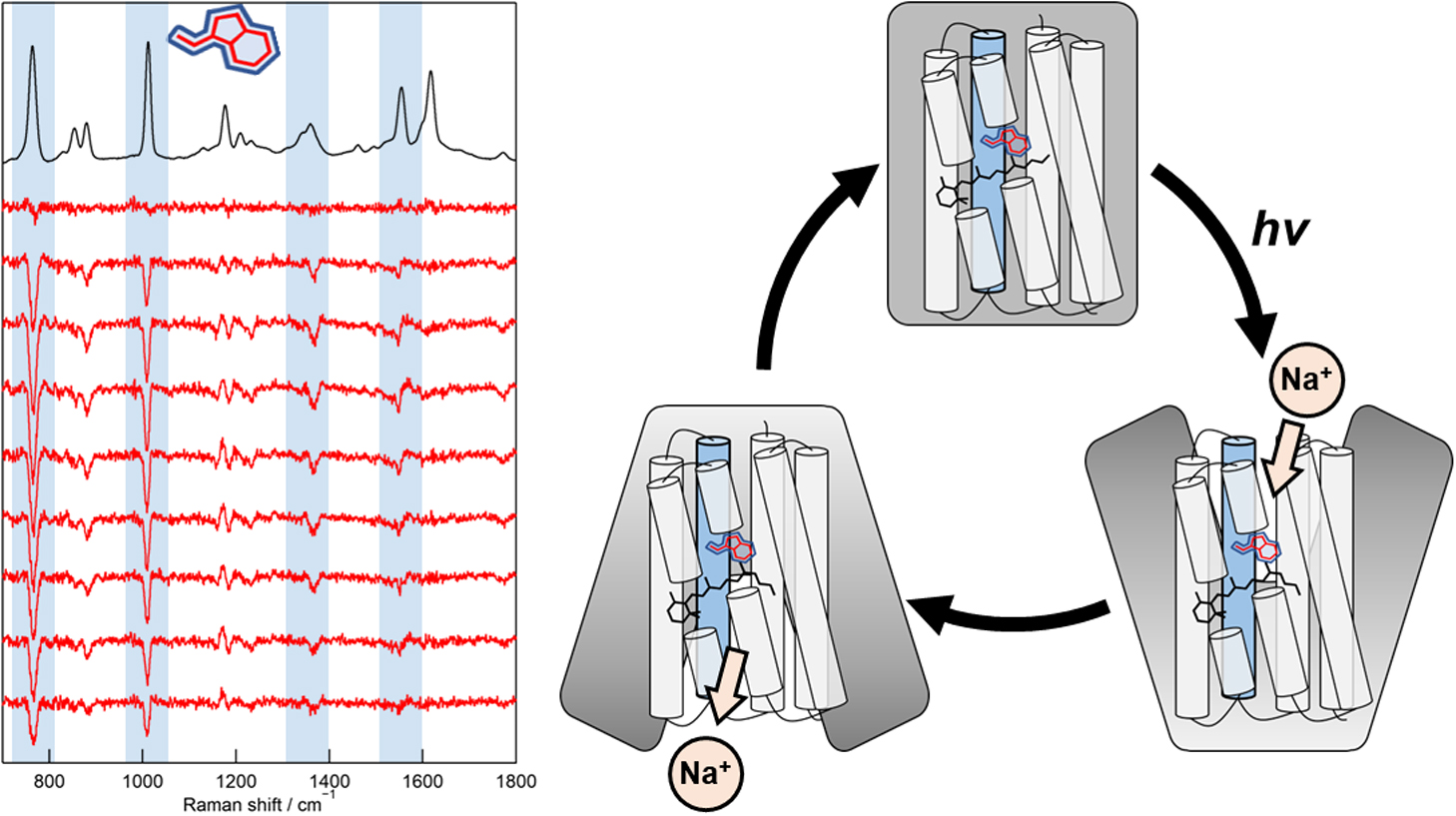
- 著者
- Akihiro Otomo Misao Mizuno Keiichi Inoue Hideki Kandori Yasuhisa Mizutani
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.e201016, (Released:2023-02-25)
Direct observation of protein structural changes during ion transport in ion pumps provides valuable insights into the mechanism of ion transport. In this study, we examined structural changes in the light-driven sodium ion (Na+) pump rhodopsin KR2 on the sub-millisecond time scale, corresponding with the uptake and release of Na+. We compared the ion-pumping activities and transient absorption spectra of WT and the W215F mutant, in which the Trp215 residue located near the retinal chromophore on the cytoplasmic side was replaced with a Phe residue. Our findings indicated that atomic contacts between the bulky side chain of Trp215 and the C20 methyl group of the retinal chromophore promote relaxation of the retinal chromophore from the 13-cis to the all-trans form. Since Trp215 is conserved in other ion-pumping rhodopsins, the present results suggest that this residue commonly acts as a mechanical transducer. In addition, we measured time-resolved ultraviolet resonance Raman (UVRR) spectra to show that the environment around Trp215 becomes less hydrophobic at 1 ms after photoirradiation and recovers to the unphotolyzed state with a time constant of around 10 ms. These time scales correspond to Na+ uptake and release, suggesting evolution of a transient ion channel at the cytoplasmic side for Na+ uptake, consistent with the alternating-access model of ion pumps. The time-resolved UVRR technique has potential for application to other ion-pumping rhodopsins and could provide further insights into the mechanism of ion transport.
- 著者
- Taro Furubayashi Norikazu Ichihashi
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.19, pp.e190005, 2022 (Released:2022-02-26)
- 参考文献数
- 43
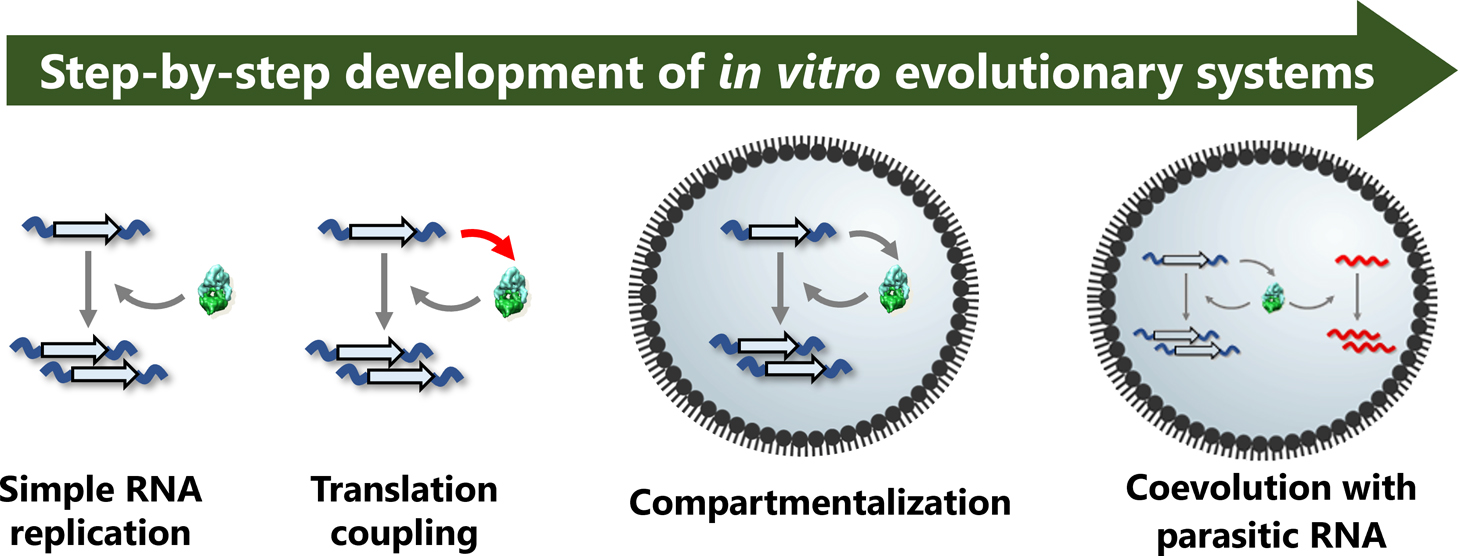
How can evolution assemble lifeless molecules into a complex living organism? The emergent process of biological complexity in the origin of life is a big mystery in biology. In vitro evolution of artificial molecular replication systems offers unique experimental opportunities to probe possible pathways of a simple molecular system approaching a complex life-like system. This review focuses on experimental efforts to examine evolvability of molecules in vitro from the pioneering Spiegelman’s experiment to our latest research on an artificial RNA self-replication system. Genetic translation and compartmentalization are shown to enable sustainable replication and evolution. Latest studies are revealing that coevolution of self-replicating “host replicators” and freeloading “parasitic replicators” is crucial to extend evolvability of a molecular replication system for continuous evolution and emergence of diversity. Intense competition between hosts and parasites would have existed even before the origin of life and contributed to generating complex molecular ecosystems. This review article is an extended version of the Japanese article “An in vitro evolutionary journey of an artificial RNA replication system towards biological complexity” published in SEIBUTSU-BUTSURI Vol.61, p.240–244 (2021).”
9 0 0 0 OA My various thoughts on actin
- 著者
- Fumio Oosawa
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.15, pp.151-158, 2018 (Released:2018-07-18)
- 参考文献数
- 42
- 被引用文献数
- 1 3
An enormous amount of research has been performed to characterize actin dynamics. Structural biology investigations have determined the localization of main chains and their changes coupled with G (Globular)-F (Filamentous) transformation of actin, whereas local thermal fluctuations that may be caused by free rotations of the tips of side chains are not yet fully investigated. This paper argues if the entropy change of actin accompanied by the G-F transformation is simply attributable to the changes in hydration. It took almost 10 years to understand that the actin filament is semi-flexible. This flexibility was visually confirmed as the development of optical microscope techniques, and the direct observation of actin severing events in the presence of actin binding proteins became possible. Finally, I expect the deep understanding of actin dynamics will lead to the elucidation of self-assembly mechanisms of the living creature.
- 著者
- Shingo Wakao Noriko Saitoh Akinori Awazu
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.e200020, (Released:2023-04-27)
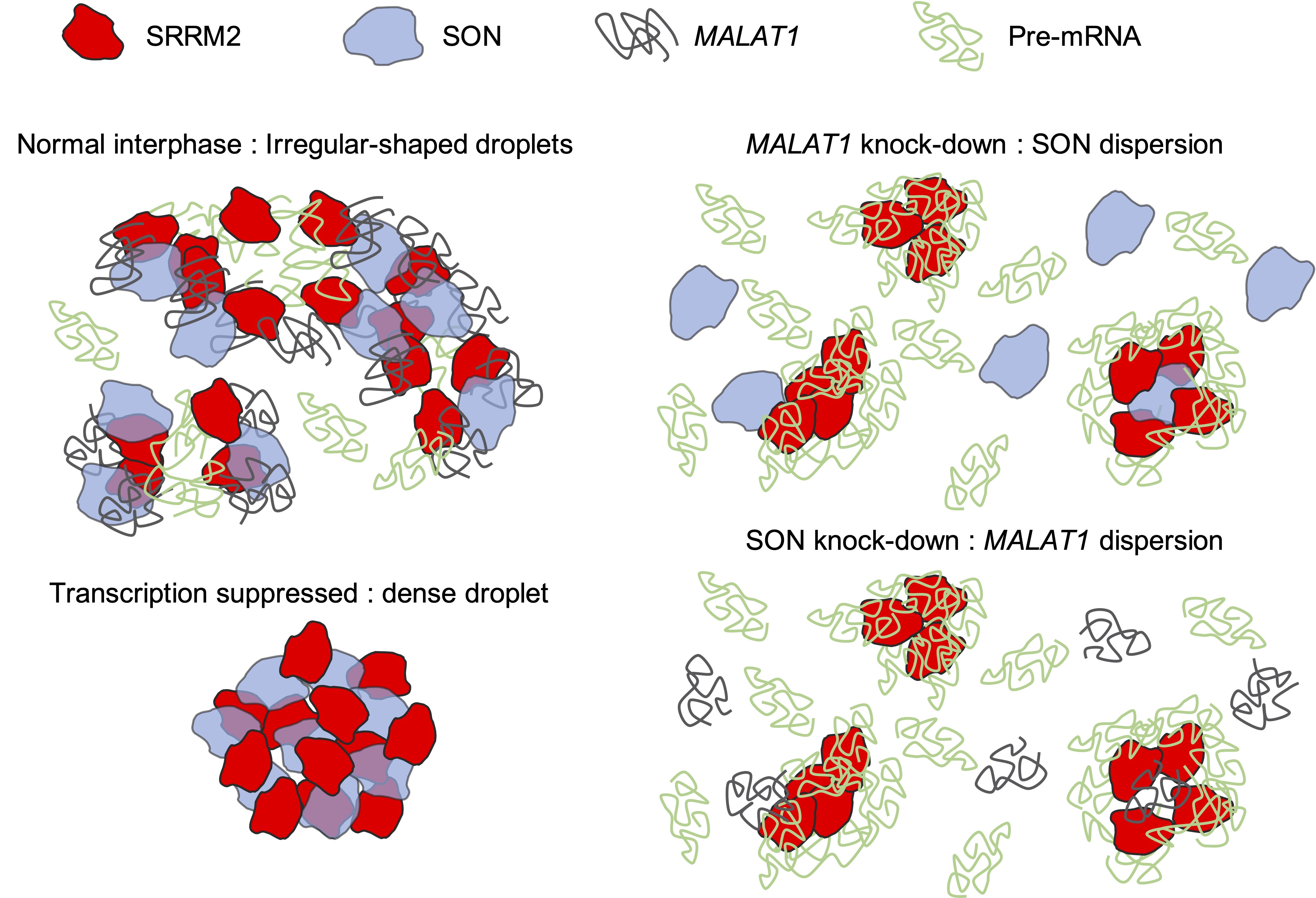
Nuclear speckles are nuclear bodies consisting of populations of small and irregularly shaped droplet-like molecular condensates that contain various splicing factors. Recent experiments have revealed the following structural features of nuclear speckles: (I) Each molecular condensate contains SON and SRRM2 proteins, and MALAT1 non-coding RNA surrounds these condensates; (II) During normal interphase of the cell cycle in multicellular organisms, these condensates are broadly distributed throughout the nucleus. In contrast, when cell transcription is suppressed, the condensates fuse and form strongly condensed spherical droplets; (III) SON is dispersed spatially in MALAT1 knocked-down cells and MALAT1 is dispersed in SON knocked-down cells because of the collapse of the nuclear speckles. However, the detailed interactions among the molecules that are mechanistically responsible for the structural variation remain unknown. In this study, a coarse-grained molecular dynamics model of the nuclear speckle was developed by considering the dynamics of SON, SRRM2, MALAT1, and pre-mRNA as representative components of the condensates. The simulations reproduced the structural changes, which were used to predict the interaction network among the representative components of the condensates.
8 0 0 0 OA Dieter Oesterhelt (1940–2022): Life with light and color, pioneer of membrane protein research
- 著者
- Peter Hegemann Hartmut Michel
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.e201010, (Released:2023-01-26)
- 被引用文献数
- 1
- 著者
- Kento Yonezawa Masatsuyo Takahashi Keiko Yatabe Yasuko Nagatani Nobutaka Shimizu
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.e200001, (Released:2023-01-07)
- 被引用文献数
- 7
Recent small-angle X-ray scattering (SAXS) for biological macromolecules (BioSAXS) is generally combined with size-exclusion chromatography (SEC-SAXS) at synchrotron facilities worldwide. For SEC-SAXS analysis, the final scattering profile for the target molecule is calculated from a large volume of continuously collected data. It would be ideal to automate this process; however, several complex problems exist regarding data measurement and analysis that have prevented automation. Here, we developed the analytical software MOLASS (Matrix Optimization with Low-rank factorization for Automated analysis of SEC-SAXS) to automatically calculate the final scattering profiles for solution structure analysis of target molecules. In this paper, the strategies for automatic analysis of SEC-SAXS data are described, including correction of baseline-drift using a low percentile method, optimization of peak decompositions composed of multiple scattering components using modified Gaussian fitting against the chromatogram, and rank determination for extrapolation to infinite dilution. In order to easily calculate each scattering component, the Moore-Penrose pseudo-inverse matrix is adopted as a basic calculation. Furthermore, this analysis method, in combination with UV–visible spectroscopy, led to better results in terms of accuracy in peak decomposition. Therefore, MOLASS will be able to smoothly suggest to users an accurate scattering profile for the subsequent structural analysis.
- 著者
- Hideaki Yoshimura
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.e190007, (Released:2022-03-11)
- 著者
- Takashi Kikukawa
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.18, pp.317-326, 2021 (Released:2022-01-08)
- 参考文献数
- 48
- 被引用文献数
- 2
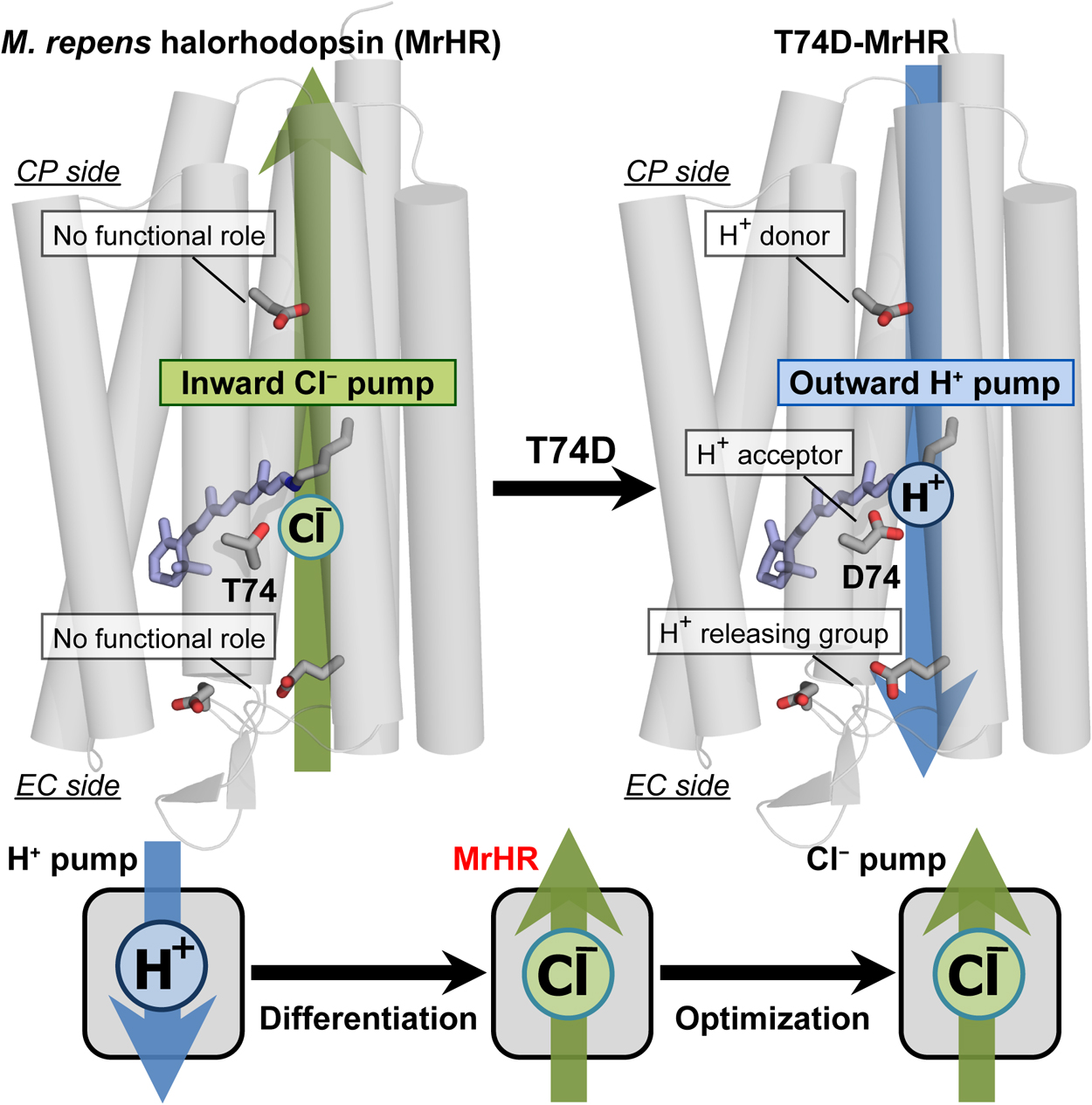
Microbial rhodopsin is a ubiquitous membrane protein in unicellular microorganisms. Similar to animal rhodopsin, this protein consists of seven transmembrane helices and the chromophore retinal. However, unlike animal rhodopsin, microbial rhodopsin acts as not only a photosignal receptor but also a light-activated ion transporter and light-switchable enzyme. In this article, the third Cl– pump microbial rhodopsin will be introduced. The physiological importance of Cl– pumps has not been clarified. Despite this, their mechanisms, especially that of the first Cl– pump halorhodopsin (HR), have been studied to characterize them as model proteins for membrane anion transporters. The third Cl– pump defines a phylogenetic cluster distinct from other microbial rhodopsins. However, this Cl– pump conserves characteristic residues for not only the Cl– pump HR but also the H+ pump bacteriorhodopsin (BR). Reflecting close similarity to BR, the third Cl– pump begins to pump H+ outwardly after single amino acid replacement. This mutation activates several residues that have no roles in the original Cl– pump function but act as important H+ relay residues in the H+ pump mutant. Thus, the third Cl– pump might be the model protein for functional differentiation because this rhodopsin seems to be the Cl– pump occurring immediately after functional differentiation from the BR-type H+ pump.
- 著者
- Takayoshi Tomono Hisao Kojima Satoshi Fukuchi Yukako Tohsato Masahiro Ito
- 出版者
- 一般社団法人 日本生物物理学会
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.12, pp.57-68, 2015 (Released:2015-11-12)
- 参考文献数
- 59
- 被引用文献数
- 5
Glycans play important roles in such cell-cell interactions as signaling and adhesion, including processes involved in pathogenic infections, cancers, and neurological diseases. Glycans are biosynthesized by multiple glycosyltransferases (GTs), which function sequentially. Excluding mucin-type O-glycosylation, the non-reducing terminus of glycans is biosynthesized in the Golgi apparatus after the reducing terminus is biosynthesized in the ER. In the present study, we performed genome-wide analyses of human GTs by investigating the degree of conservation of homologues in other organisms, as well as by elucidating the phylogenetic relationship between cephalochordates and urochordates, which has long been controversial in deuterostome phylogeny. We analyzed 173 human GTs and functionally linked glycan synthesis enzymes by phylogenetic profiling and clustering, compiled orthologous genes from the genomes of other organisms, and converted them into a binary sequence based on the presence (1) or absence (0) of orthologous genes in the genomes. Our results suggest that the non-reducing terminus of glycans is biosynthesized by newly evolved GTs. According to our analysis, the phylogenetic profiles of GTs resemble the phylogenetic tree of life, where deuterostomes, metazoans, and eukaryotes are resolved into separate branches. Lineage-specific GTs appear to play essential roles in the divergence of these particular lineages. We suggest that urochordates lose several genes that are conserved among metazoans, such as those expressing sialyltransferases, and that the Golgi apparatus acquires the ability to synthesize glycans after the ER acquires this function.