5 0 0 0 OA Specificity of broad protein interaction surfaces for proteins with multiple binding partners
- 著者
- Nobuyuki Uchikoga Yuri Matsuzaki Masahito Ohue Yutaka Akiyama
- 出版者
- 一般社団法人 日本生物物理学会
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.13, pp.105-115, 2016 (Released:2016-07-14)
- 参考文献数
- 31
- 被引用文献数
- 7
Analysis of protein-protein interaction networks has revealed the presence of proteins with multiple interaction ligand proteins, such as hub proteins. For such proteins, multiple ligands would be predicted as interacting partners when predicting all-to-all protein-protein interactions (PPIs). In this work, to obtain a better understanding of PPI mechanisms, we focused on protein interaction surfaces, which differ between protein pairs. We then performed rigid-body docking to obtain information of interfaces of a set of decoy structures, which include many possible interaction surfaces between a certain protein pair. Then, we investigated the specificity of sets of decoy interactions between true binding partners in each case of alpha-chymotrypsin, actin, and cyclin-dependent kinase 2 as test proteins having multiple true binding partners. To observe differences in interaction surfaces of docking decoys, we introduced broad interaction profiles (BIPs), generated by assembling interaction profiles of decoys for each protein pair. After cluster analysis, the specificity of BIPs of true binding partners was observed for each receptor. We used two types of BIPs: those involved in amino acid sequences (BIP-seqs) and those involved in the compositions of interacting amino acid residue pairs (BIP-AAs). The specificity of a BIP was defined as the number of group members including all true binding partners. We found that BIP-AA cases were more specific than BIP-seq cases. These results indicated that the composition of interacting amino acid residue pairs was sufficient for determining the properties of protein interaction surfaces.
- 著者
- MASAHITO OHUE YURI MATSUZAKI YUTAKA AKIYAMA
- 出版者
- Japanese Society for Bioinformatics
- 雑誌
- Genome Informatics (ISSN:09199454)
- 巻号頁・発行日
- vol.25, no.1, pp.25-39, 2011 (Released:2011-08-03)
- 参考文献数
- 22
Elucidating protein-RNA interactions (PRIs) is important for understanding many cellular systems. We developed a PRI prediction method by using a rigid-body protein-RNA docking calculation with tertiary structure data. We evaluated this method by using 78 protein-RNA complex structures from the Protein Data Bank. We predicted the interactions for pairs in 78×78 combinations. Of these, 78 original complexes were defined as positive pairs, and the other 6,006 complexes were defined as negative pairs; then an F-measure value of 0.465 was obtained with our prediction system.
- 著者
- Shumpei Matsuno Masahito Ohue Yutaka Akiyama
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.17, pp.2-13, 2020 (Released:2020-02-07)
- 参考文献数
- 30
- 被引用文献数
- 3
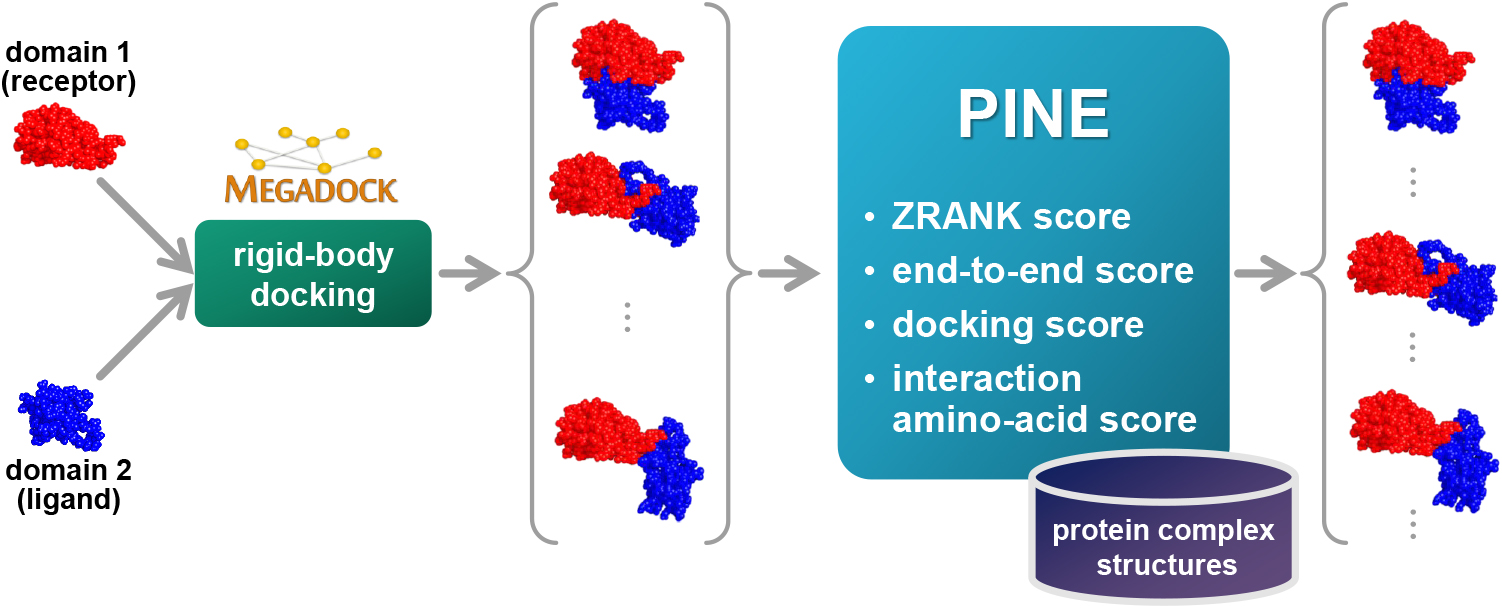
Protein functions can be predicted based on their three-dimensional structures. However, many multidomain proteins have unstable structures, making it difficult to determine the whole structure in biological experiments. Additionally, multidomain proteins are often decomposed and identified based on their domains, with the structure of each domain often found in public databases. Recent studies have advanced structure prediction methods of multidomain proteins through computational analysis. In existing methods, proteins that serve as templates are used for three-dimensional structure prediction. However, when no protein template is available, the accuracy of the prediction is decreased. This study was conducted to predict the structures of multidomain proteins without the need for whole structure templates.We improved structure prediction methods by performing rigid-body docking from the structure of each domain and reranking a structure closer to the correct structure to have a higher value. In the proposed method, the score for the domain-domain interaction obtained without a structural template of the multidomain protein and score for the three-dimensional structure obtained during docking calculation were newly incorporated into the score function. We successfully predicted the structures of 50 of 55 multidomain proteins examined in the test dataset.Interaction residue pair information of the protein-protein complex interface contributes to domain reorganizations even when a structural template for a multidomain protein cannot be obtained. This approach may be useful for predicting the structures of multidomain proteins with important biochemical functions.
- 著者
- Chaojie Zhang Koichi Shirahata Shuji Suzuki Yutaka Akiyama Satoshi Matsuoka
- 雑誌
- 研究報告計算機アーキテクチャ(ARC)
- 巻号頁・発行日
- vol.2014-ARC-213, no.29, pp.1-7, 2014-12-02
Homology search to be used in emerging bioinformatics problems such as metagenomics is of increasing importance and challenge as its application area grows more broadly while the computational complexity is increasing, thus requiring massive parallel data processing. Earlier work by some of the authors have devised novel algorithms such as GHOSTX, but the master-worker parallelization to enumerate and schedule for data processing was done with a privately developed, MPI-based master-worker framework called GHOST-MP. An alternative is to utilize the now-popular big data software substrates, such as MapReduce with abundant associated software tool-chains, but it is not clear whether the massive resource required by metagenomic homology search would not overwhelm its known limitations. By converting the GHOST-MP master-worker data processing pipeline to accommodate MapReduce, and benchmarking them on a variety of high-performance MapReduce incarnations including Hadoop and Spark, we attempt to characterize the appropriateness of MapReduce as a generic framework for metagenomics that embody extremely resource consuming requirements for both compute and data.
- 著者
- Wisnu Ananta Kusuma Takashi Ishida Yutaka Akiyama
- 出版者
- Information Processing Society of Japan
- 雑誌
- IPSJ Transactions on Bioinformatics (ISSN:18826679)
- 巻号頁・発行日
- vol.4, pp.21-33, 2011 (Released:2011-11-04)
- 参考文献数
- 27
- 被引用文献数
- 2 2
De novo DNA sequence assembly is very important in genome sequence analysis. In this paper, we investigated two of the major approaches for de novo DNA sequence assembly of very short reads: overlap-layout-consensus (OLC) and Eulerian path. From that investigation, we developed a new assembly technique by combining the OLC and the Eulerian path methods in a hierarchical process. The contigs yielded by these two approaches were treated as reads and were assembled again to yield longer contigs. We tested our approach using three real very-short-read datasets generated by an Illumina Genome Analyzer and four simulated very-short-read datasets that contained sequencing errors. The sequencing errors were modeled based on Illumina's sequencing technology. As a result, our combined approach yielded longer contigs than those of Edena (OLC) and Velvet (Eulerian path) in various coverage depths and was comparable to SOAPdenovo, in terms of N50 size and maximum contig lengths. The assembly results were also validated by comparing contigs that were produced by assemblers with their reference sequence from an NCBI database. The results show that our approach produces more accurate results than Velvet, Edena, and SOAPdenovo alone. This comparison indicates that our approach is a viable way to assemble very short reads from next generation sequencers.
- 著者
- Keisuke Yanagisawa Takashi Ishida Yutaka Akiyama
- 出版者
- 一般社団法人 情報処理学会
- 雑誌
- IPSJ Transactions on Bioinformatics (ISSN:18826679)
- 巻号頁・発行日
- vol.8, pp.21-27, 2015 (Released:2015-08-19)
- 参考文献数
- 21
It is necessary to confirm that a new drug can be appropriately cleared from the human body. However, checking the clearance pathway of a drug in the human body requires clinical trials, and therefore requires large cost. Thus, computational methods for drug clearance pathway prediction have been studied. The proposed prediction methods developed previously were based on a supervised learning algorithm, which requires clearance pathway information for all drugs in a training set as input labels. However, these data are often insufficient in its numbers because of the high cost of their acquisition. In this paper, we propose a new drug clearance pathway prediction method based on semi-supervised learning, which can use not only labeled data but also unlabeled data. We evaluated the effectiveness of our method, focusing on the cytochrome P450 2C19 enzyme, which is involved in one of the major clearance pathways.