- 著者
- Hiroaki Hata Duy Phuoc Tran Mohamed Marzouk Sobeh Akio Kitao
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.bppb-v18.037, (Released:2021-12-04)
- 被引用文献数
- 22
- 著者
- Junko Taguchi Akio Kitao
- 出版者
- 一般社団法人 日本生物物理学会
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.13, pp.117-126, 2016 (Released:2016-07-14)
- 参考文献数
- 37
- 被引用文献数
- 7
We examine the dynamic features of non-trivial allosteric binding sites to elucidate potential drug binding sites. These allosteric sites were previously found to be allosteric after determination of the protein-drug co-crystal structure. After comprehensive search in the Protein Data Bank, we identify 10 complex structures with allosteric ligands whose structures are very similar to their functional forms. Then, possible pockets on the protein surface are searched as potential ligand binding sites. To mimic ligand binding to the pocket, complex models are generated to fill out each pocket with pseudo ligand blocks consisting of spheres. Normal mode analysis of the elastic network model is performed for the complex models and unbound structures to assess the change of protein dynamics induced by ligand binding. We examine nine profiles to describe the dynamic and positional characteristics of the pockets, and identify the change of fluctuation around the ligand, ΔMSFbs, as the best profile for distinguishing the allosteric sites from the other sites in 8 structures. These cases should be considered as examples of dynamics-driven allostery, which accompanies significant changes in protein dynamics. ΔMSFbs is suggested to be used for the search of potential dynamics-driven allosteric sites in proteins for drug discovery.
- 著者
- Yasumasa Joti Akio Kitao
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.16, pp.240-247, 2019 (Released:2019-11-29)
- 参考文献数
- 28
- 被引用文献数
- 2
Terahertz time-domain spectra (THz-TDS) were investigated using the results of molecular dynamics (MD) simulations of Staphylococcal nuclease at two hydration states in the temperature range between 100 and 300 K. The temperature dependence of THz-TDS was found to differ significantly from that of the incoherent neutron scattering spectra (INSS) calculated from the same MD simulation results. We further examined contributions of the mutual and auto-correlations of the atomic fluctuations to THz-TDS and found that the negative value of the former contribution nearly canceled out the positive value of the latter, resulting in a monotonic increase of the reduced absorption cross section. Because of this cancellation, no distinct broad peak was observed in the absorption lineshape function of THz-TDS, whereas the protein boson peak was observed in INSS. The contribution of water molecules to THz-TDS was extremely large for the hydrated protein at temperatures above 200 K, in which large-amplitude motions of water were excited. The combination of THz-TDS, INSS and MD simulations has the potential to extract function-relevant protein dynamics occurring on the picosecond to nanosecond timescale.
- 著者
- Kazuhiro Takemura Akio Kitao
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.16, pp.295-303, 2019 (Released:2019-11-29)
- 参考文献数
- 36
- 被引用文献数
- 4
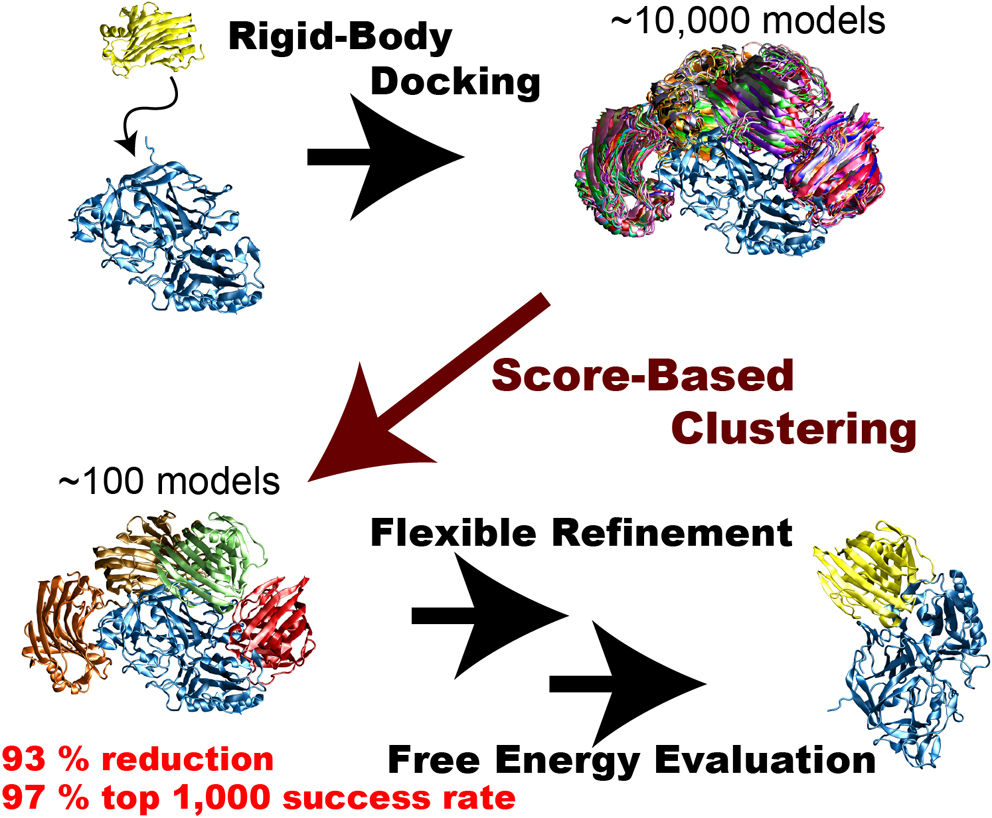
Rigid-body protein-protein docking is very efficient in generating tens of thousands of docked complex models (decoys) in a very short time without considering structure change upon binding, but typical docking scoring functions are not necessarily sufficiently accurate to narrow these decoys down to a small number of plausible candidates. Flexible refinements and sophisticated evaluation of the decoys are thus required to achieve more accurate prediction. Since this process is time-consuming, an efficient screening method to reduce the number of decoys is necessary immediately following rigid-body dockings. We attempted to develop an efficient screening method by clustering decoys generated by the rigid-body docking ZDOCK. We introduced the three metrics ligand-root-mean-square deviation (L-RMSD), interface-ligand-RMSD (iL-RMSD), and the fraction of common contacts (FCC), and examined various ranges of cut-offs for clusters to determine the best set of clustering parameters. Although the employed clustering algorithm is simple, it successfully reduced the number of decoys. Using iL-RMSD with a cut-off radius of 8 Å, the number of decoys that contain at least one near-native model with 90% probability decreased from 4,808 to 320, a 93% reduction in the original number of decoys. Using FCC for the clustering step, the top 1,000 success rates, defined as the probability that the top 1,000 models contain at least one near-native structure, reached 97%. We conclude that the proposed method is very efficient in selecting a small number of decoys that include near-native decoys.
- 著者
- Hiroaki Hata Duy Phuoc Tran Mohamed Marzouk Sobeh Akio Kitao
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.18, pp.305-316, 2021 (Released:2021-12-22)
- 参考文献数
- 68
- 被引用文献数
- 22
We recently proposed a computational procedure to simulate the dissociation of protein/ligand complexes using the dissociation Parallel Cascade Selection Molecular Dynamics simulation (dPaCS-MD) method and to analyze the generated trajectories using the Markov state model (MSM). This procedure, called dPaCS-MD/MSM, enables calculation of the dissociation free energy profile and the standard binding free energy. To examine whether this method can reproduce experimentally determined binding free energies for a variety of systems, we used it to investigate the dissociation of three protein/ligand complexes: trypsin/benzamine, FKBP/FK506, and adenosine A2A receptor/T4E. First, dPaCS-MD generated multiple dissociation pathways within a reasonable computational time for all the complexes, although the complexes differed significantly in the size of the molecules and in intermolecular interactions. Subsequent MSM analyses produced free energy profiles for the dissociations, which provided insights into how each ligand dissociates from the protein. The standard binding free energies obtained by dPaCS-MD/MSM are in good agreement with experimental values for all the complexes. We conclude that dPaCS-MD/MSM can accurately calculate the binding free energies of these complexes.