33 0 0 0 OA Light-driven Proton Pumps as a Potential Regulator for Carbon Fixation in Marine Diatoms
- 著者
- Susumu Yoshizawa Tomonori Azuma Keiichi Kojima Keisuke Inomura Masumi Hasegawa Yosuke Nishimura Masuzu Kikuchi Gabrielle Armin Yuya Tsukamoto Hideaki Miyashita Kentaro Ifuku Takashi Yamano Adrian Marchetti Hideya Fukuzawa Yuki Sudo Ryoma Kamikawa
- 出版者
- Japanese Society of Microbial Ecology / Japanese Society of Soil Microbiology / Taiwan Society of Microbial Ecology / Japanese Society of Plant Microbe Interactions / Japanese Society for Extremophiles
- 雑誌
- Microbes and Environments (ISSN:13426311)
- 巻号頁・発行日
- vol.38, no.2, pp.ME23015, 2023 (Released:2023-06-20)
- 参考文献数
- 46
- 被引用文献数
- 4
Diatoms are a major phytoplankton group responsible for approximately 20% of carbon fixation on Earth. They perform photosynthesis using light-harvesting chlorophylls located in plastids, an organelle obtained through eukaryote-eukaryote endosymbiosis. Microbial rhodopsin, a photoreceptor distinct from chlorophyll-based photosystems, was recently identified in some diatoms. However, the physiological function of diatom rhodopsin remains unclear. Heterologous expression techniques were herein used to investigate the protein function and subcellular localization of diatom rhodopsin. We demonstrated that diatom rhodopsin acts as a light-driven proton pump and localizes primarily to the outermost membrane of four membrane-bound complex plastids. Using model simulations, we also examined the effects of pH changes inside the plastid due to rhodopsin-mediated proton transport on photosynthesis. The results obtained suggested the involvement of rhodopsin-mediated local pH changes in a photosynthetic CO2-concentrating mechanism in rhodopsin-possessing diatoms.
- 著者
- Yu Nakajima Takashi Tsukamoto Yohei Kumagai Yoshitoshi Ogura Tetsuya Hayashi Jaeho Song Takashi Kikukawa Makoto Demura Kazuhiro Kogure Yuki Sudo Susumu Yoshizawa
- 出版者
- Japanese Society of Microbial Ecology · The Japanese Society of Soil Microbiology
- 雑誌
- Microbes and Environments (ISSN:13426311)
- 巻号頁・発行日
- vol.33, no.1, pp.89-97, 2018 (Released:2018-03-29)
- 参考文献数
- 43
- 被引用文献数
- 17
Light-driven ion-pumping rhodopsins are widely distributed among bacteria, archaea, and eukaryotes in the euphotic zone of the aquatic environment. H+-pumping rhodopsin (proteorhodopsin: PR), Na+-pumping rhodopsin (NaR), and Cl−-pumping rhodopsin (ClR) have been found in marine bacteria, which suggests that these genes evolved independently in the ocean. Putative microbial rhodopsin genes were identified in the genome sequences of marine Cytophagia. In the present study, one of these genes was heterologously expressed in Escherichia coli cells and the rhodopsin protein named Rubricoccus marinus halorhodopsin (RmHR) was identified as a light-driven inward Cl− pump. Spectroscopic assays showed that the estimated dissociation constant (Kd,int.) of this rhodopsin was similar to that of haloarchaeal halorhodopsin (HR), while the Cl−-transporting photoreaction mechanism of this rhodopsin was similar to that of HR, but different to that of the already-known marine bacterial ClR. This amino acid sequence similarity also suggested that this rhodopsin is similar to haloarchaeal HR and cyanobacterial HRs (e.g., SyHR and MrHR). Additionally, a phylogenetic analysis revealed that retinal biosynthesis pathway genes (blh and crtY) belong to a phylogenetic lineage of haloarchaea, indicating that these marine Cytophagia acquired rhodopsin-related genes from haloarchaea by lateral gene transfer. Based on these results, we concluded that inward Cl−-pumping rhodopsin is present in genera of the class Cytophagia and may have the same evolutionary origins as haloarchaeal HR.
18 0 0 0 OA Bacterium Lacking a Known Gene for Retinal Biosynthesis Constructs Functional Rhodopsins
- 著者
- Yu Nakajima Keiichi Kojima Yuichiro Kashiyama Satoko Doi Ryosuke Nakai Yuki Sudo Kazuhiro Kogure Susumu Yoshizawa
- 出版者
- Japanese Society of Microbial Ecology / Japanese Society of Soil Microbiology / Taiwan Society of Microbial Ecology / Japanese Society of Plant Microbe Interactions / Japanese Society for Extremophiles
- 雑誌
- Microbes and Environments (ISSN:13426311)
- 巻号頁・発行日
- vol.35, no.4, pp.ME20085, 2020 (Released:2020-12-05)
- 参考文献数
- 36
- 被引用文献数
- 15
Microbial rhodopsins, comprising a protein moiety (rhodopsin apoprotein) bound to the light-absorbing chromophore retinal, function as ion pumps, ion channels, or light sensors. However, recent genomic and metagenomic surveys showed that some rhodopsin-possessing prokaryotes lack the known genes for retinal biosynthesis. Since rhodopsin apoproteins cannot absorb light energy, rhodopsins produced by prokaryotic strains lacking genes for retinal biosynthesis are hypothesized to be non-functional in cells. In the present study, we investigated whether Aurantimicrobium minutum KNCT, which is widely distributed in terrestrial environments and lacks any previously identified retinal biosynthesis genes, possesses functional rhodopsin. We initially measured ion transport activity in cultured cells. A light-induced pH change in a cell suspension of rhodopsin-possessing bacteria was detected in the absence of exogenous retinal. Furthermore, spectroscopic analyses of the cell lysate and HPLC-MS/MS analyses revealed that this strain contained an endogenous retinal. These results confirmed that A. minutum KNCT possesses functional rhodopsin and, hence, produces retinal via an unknown biosynthetic pathway. These results suggest that rhodopsin-possessing prokaryotes lacking known retinal biosynthesis genes also have functional rhodopsins.
- 著者
- Yu Nakajima Takashi Tsukamoto Yohei Kumagai Yoshitoshi Ogura Tetsuya Hayashi Jaeho Song Takashi Kikukawa Makoto Demura Kazuhiro Kogure Yuki Sudo Susumu Yoshizawa
- 出版者
- Japanese Society of Microbial Ecology · The Japanese Society of Soil Microbiology
- 雑誌
- Microbes and Environments (ISSN:13426311)
- 巻号頁・発行日
- pp.ME17197, (Released:2018-03-16)
- 被引用文献数
- 17
Light-driven ion-pumping rhodopsins are widely distributed among bacteria, archaea, and eukaryotes in the euphotic zone of the aquatic environment. H+-pumping rhodopsin (proteorhodopsin: PR), Na+-pumping rhodopsin (NaR), and Cl–-pumping rhodopsin (ClR) have been found in marine bacteria, which suggests that these genes evolved independently in the ocean. Putative microbial rhodopsin genes were identified in the genome sequences of marine Cytophagia. In the present study, one of these genes was heterologously expressed in Escherichia coli cells and the rhodopsin protein named Rubricoccus marinus halorhodopsin (RmHR) was identified as a light-driven inward Cl– pump. Spectroscopic assays showed that the estimated dissociation constant (Kd,int.) of this rhodopsin was similar to that of haloarchaeal halorhodopsin (HR), while the Cl–-transporting photoreaction mechanism of this rhodopsin was similar to that of HR, but different to that of the already-known marine bacterial ClR. This amino acid sequence similarity also suggested that this rhodopsin is similar to haloarchaeal HR and cyanobacterial HRs (e.g., SyHR and MrHR). Additionally, a phylogenetic analysis revealed that retinal biosynthesis pathway genes (blh and crtY) belong to a phylogenetic lineage of haloarchaea, indicating that these marine Cytophagia acquired rhodopsin-related genes from haloarchaea by lateral gene transfer. Based on these results, we concluded that inward Cl–-pumping rhodopsin is present in genera of the class Cytophagia and may have the same evolutionary origins as haloarchaeal HR.
- 著者
- Marie Kurihara Vera Thiel Hirona Takahashi Keiichi Kojima David M. Ward Donald A. Bryant Makoto Sakai Susumu Yoshizawa Yuki Sudo
- 出版者
- The Pharmaceutical Society of Japan
- 雑誌
- Chemical and Pharmaceutical Bulletin (ISSN:00092363)
- 巻号頁・発行日
- vol.71, no.2, pp.154-164, 2023-02-01 (Released:2023-02-01)
- 参考文献数
- 45
- 被引用文献数
- 1
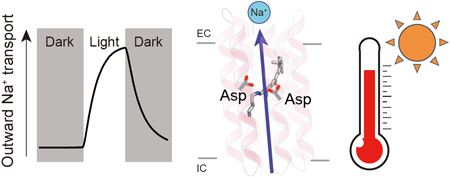
Rhodopsins are transmembrane proteins with retinal chromophores that are involved in photo-energy conversion and photo-signal transduction in diverse organisms. In this study, we newly identified and characterized a rhodopsin from a thermophilic bacterium, Bellilinea sp. Recombinant Escherichia coli cells expressing the rhodopsin showed light-induced alkalization of the medium only in the presence of sodium ions (Na+), and the alkalization signal was enhanced by addition of a protonophore, indicating an outward Na+ pump function across the cellular membrane. Thus, we named the protein Bellilinea Na+-pumping rhodopsin, BeNaR. Of note, its Na+-pumping activity is significantly greater than that of the known Na+-pumping rhodopsin, KR2. We further characterized its photochemical properties as follows: (i) Visible spectroscopy and HPLC revealed that BeNaR has an absorption maximum at 524 nm with predominantly (>96%) the all-trans retinal conformer. (ii) Time-dependent thermal denaturation experiments revealed that BeNaR showed high thermal stability. (iii) The time-resolved flash-photolysis in the nanosecond to millisecond time domains revealed the presence of four kinetically distinctive photointermediates, K, L, M and O. (iv) Mutational analysis revealed that Asp101, which acts as a counterion, and Asp230 around the retinal were essential for the Na+-pumping activity. From the results, we propose a model for the outward Na+-pumping mechanism of BeNaR. The efficient Na+-pumping activity of BeNaR and its high stability make it a useful model both for ion transporters and optogenetics tools.
- 著者
- Md Mehedi Iqbal Masahiko Nishimura Md. Nurul Haider Masayoshi Sano Minoru Ijichi Kazuhiro Kogure Susumu Yoshizawa
- 出版者
- Japanese Society of Microbial Ecology / Japanese Society of Soil Microbiology / Taiwan Society of Microbial Ecology / Japanese Society of Plant Microbe Interactions / Japanese Society for Extremophiles
- 雑誌
- Microbes and Environments (ISSN:13426311)
- 巻号頁・発行日
- vol.36, no.4, pp.ME21037, 2021 (Released:2021-10-13)
- 参考文献数
- 81
- 被引用文献数
- 10
Zostera marina (eelgrass) is a widespread seagrass species that forms diverse and productive habitats along coast lines throughout much of the northern hemisphere. The present study investigated the microbial consortia of Z. marina growing at Futtsu clam-digging beach, Chiba prefecture, Japan. The following environmental samples were collected: sediment, seawater, plant leaves, and the root-rhizome. Sediment and seawater samples were obtained from three sampling points: inside, outside, and at the marginal point of the eelgrass bed. The microbial composition of each sample was analyzed using 16S ribosomal gene amplicon sequencing. Microbial communities on the dead (withered) leaf surface markedly differed from those in sediment, but were similar to those in seawater. Eelgrass leaves and surrounding seawater were dominated by the bacterial taxa Rhodobacterales (Alphaproteobacteria), whereas Rhodobacterales were a minor group in eelgrass sediment. Additionally, we speculated that the order Sphingomonadales (Alphaproteobacteria) acts as a major degrader during the decomposition process and constantly degrades eelgrass leaves, which then spread into the surrounding seawater. Withered eelgrass leaves did not accumulate on the surface sediment because they were transported out of the eelgrass bed by wind and residual currents unique to the central part of Tokyo Bay.