1 0 0 0 OA Earthworm individualities when facing a conflict between turn alternation and aversive learning
- 著者
- Tadashi Nakashima Hajime Mushiake Kazuhiro Sakamoto
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.15, pp.159-164, 2018 (Released:2018-07-31)
- 参考文献数
- 33
- 被引用文献数
- 2 5
An individual’s personality develops through a combination of experiences and parental inheritance. When faced with a conflict, will an individual take an innate behavior or a learned one? In such situations, individuality will manifest itself. Here, we focused on turn alternation behavior, which is a habitual tendency to turn in the direction opposite the preceding turn, in earthworms (Eisenia fetida) and examined how this behavior is affected by an aversive stimulus. Of 10 earthworms, 3 were affected by the stimulus. Turn alternation deteriorated in two worms, one of which showed anti-turn alternation behavior, whereas the remaining worm showed an enhanced tendency toward turn alternation. Earthworms have a relatively simple nervous system. This study opens the door to investigate the neuronal basis for individuality that emerges between nature and nurture.
1 0 0 0 OA Light-induced difference FTIR spectroscopy of primate blue-sensitive visual pigment at 163 K
- 著者
- Shunpei Hanai Kota Katayama Hiroo Imai Hideki Kandori
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.bppb-v18.005, (Released:2021-02-13)
- 被引用文献数
- 3
1 0 0 0 OA Biphasic spatiotemporal regulation of GRB2 dynamics by p52SHC for transient RAS activation
- 著者
- Ryo Yoshizawa Nobuhisa Umeki Akihiro Yamamoto Masayuki Murata Yasushi Sako
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.18, pp.1-12, 2021 (Released:2021-02-05)
- 参考文献数
- 47
- 被引用文献数
- 7
RTK-RAS-MAPK systems are major signaling pathways for cell fate decisions. Among the several RTK species, it is known that the transient activation of ERK (MAPK) stimulates cell proliferation, whereas its sustained activation induces cell differentiation. In both instances however, RAS activation is transient, suggesting that the strict temporal regulation of its activity is critical in normal cells. RAS on the cytoplasmic side of the plasma membrane is activated by SOS through the recruitment of GRB2/SOS complex to the RTKs that are phosphorylated after stimulation with growth factors. The adaptor protein GRB2 recognizes phospho-RTKs both directly and indirectly via another adaptor protein, SHC. We here studied the regulation of GRB2 recruitment under the SHC pathway using single-molecule imaging and fluorescence correlation spectroscopy in living cells. We stimulated MCF7 cells with a differentiation factor, heregulin, and observed the translocation, complex formation, and phosphorylation of cell signaling molecules including GRB2 and SHC. Our results suggest a biphasic regulation of the GRB2/SOS-RAS pathway by SHC: At the early stage (<10 min) of stimulation, SHC increased the amplitude of RAS activity by increasing the association sites for the GRB2/SOS complex on the plasma membrane. At the later stage however, SHC suppressed RAS activation and sequestered GRB2 molecules from the membrane through the complex formation in the cytoplasm. The latter mechanism functions additively to other mechanisms of negative feedback regulation of RAS from MEK and/or ERK to complete the transient activation dynamics of RAS.
- 著者
- Yujiro Nagasaka Shoko Hososhima Naoko Kubo Takashi Nagata Hideki Kandori Keiichi Inoue Hiromu Yawo
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.17, pp.59-70, 2020 (Released:2020-07-22)
- 参考文献数
- 51
- 被引用文献数
- 5
Microbial rhodopsin is a large family of membrane proteins having seven transmembrane helices (TM1-7) with an all-trans retinal (ATR) chromophore that is covalently bound to Lys in the TM7. The Trp residue in the middle of TM3, which is homologous to W86 of bacteriorhodopsin (BR), is highly conserved among microbial rhodopsins with various light-driven functions. However, the significance of this Trp for the ion transport function of microbial rhodopsins has long remained unknown. Here, we replaced the W163 (BR W86 counterpart) of a channelrhodopsin (ChR), C1C2/ChRWR, which is a chimera between ChR1 and 2, with a smaller aromatic residue, Phe to verify its role in the ion transport. Under whole-cell patch clamp recordings from the ND7/23 cells that were transfected with the DNA plasmid coding human codon optimized C1C2/ChRWR (hWR) or its W163F mutant (hWR-W163F), the photocurrents were evoked by a pulsatile light at 475 nm. The ion-transporting activity of hWR was strongly altered by the W163F mutation in 3 points: (1) the H+ leak at positive membrane potential (Vm) and its light-adaptation, (2) the attenuation of cation channel activity and (3) the manifestation of outward H+ pump activity. All of these results strongly suggest that W163 has a role in stabilizing the structure involved in the gating-on and -off of the cation channel, the role of “gate keeper”. We can attribute the attenuation of cation channel activity to the incomplete gating-on and the H+ leak to the incomplete gating-off.
- 著者
- Masaki Sasai George Chikenji Tomoki P. Terada
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.13, pp.281-293, 2016 (Released:2016-11-18)
- 参考文献数
- 73
- 被引用文献数
- 3 5
A simple statistical mechanical model proposed by Wako and Saitô has explained the aspects of protein folding surprisingly well. This model was systematically applied to multiple proteins by Muñoz and Eaton and has since been referred to as the Wako-Saitô-Muñoz-Eaton (WSME) model. The success of the WSME model in explaining the folding of many proteins has verified the hypothesis that the folding is dominated by native interactions, which makes the energy landscape globally biased toward native conformation. Using the WSME and other related models, Saitô emphasized the importance of the hierarchical pathway in protein folding; folding starts with the creation of contiguous segments having a native-like configuration and proceeds as growth and coalescence of these segments. The Φ-values calculated for barnase with the WSME model suggested that segments contributing to the folding nucleus are similar to the structural modules defined by the pattern of native atomic contacts. The WSME model was extended to explain folding of multi-domain proteins having a complex topology, which opened the way to comprehensively understanding the folding process of multi-domain proteins. The WSME model was also extended to describe allosteric transitions, indicating that the allosteric structural movement does not occur as a deterministic sequential change between two conformations but as a stochastic diffusive motion over the dynamically changing energy landscape. Statistical mechanical viewpoint on folding, as highlighted by the WSME model, has been renovated in the context of modern methods and ideas, and will continue to provide insights on equilibrium and dynamical features of proteins.
- 著者
- Sumita Das Tomoki P. Terada Masaki Sasai
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.15, pp.136-150, 2018 (Released:2018-05-26)
- 参考文献数
- 47
- 被引用文献数
- 9
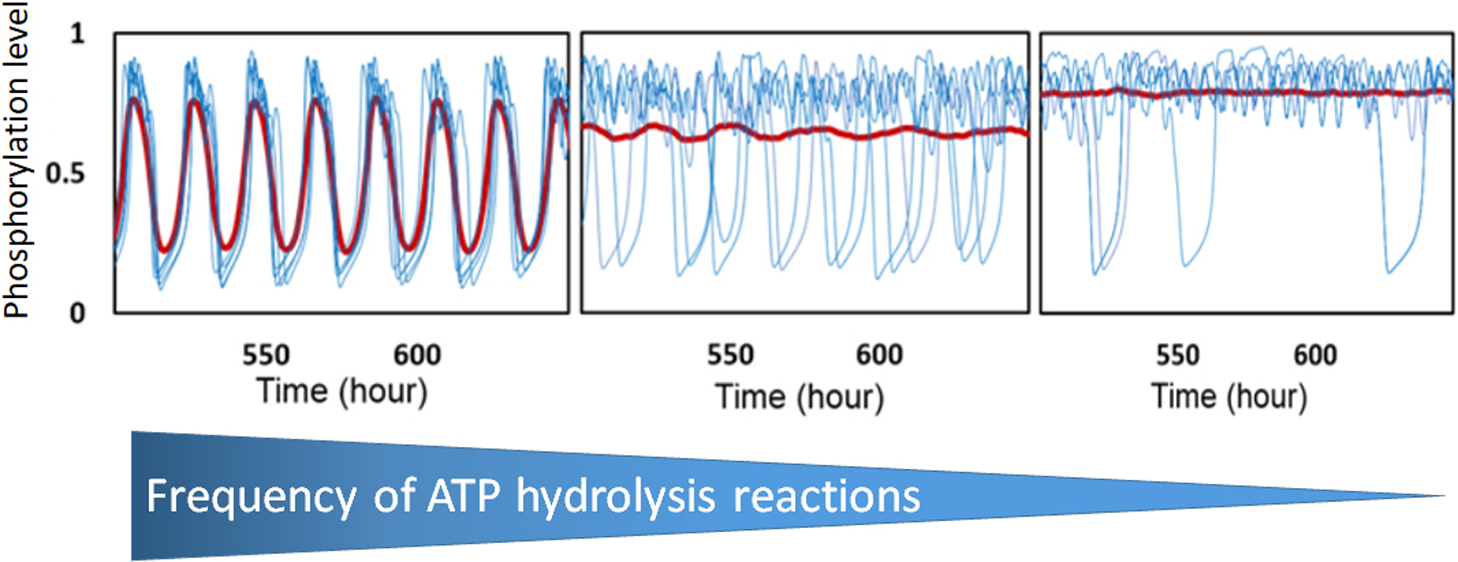
When three cyanobacterial proteins, KaiA, KaiB, and KaiC, are incubated with ATP in vitro, the phosphorylation level of KaiC hexamers shows stable oscillation with approximately 24 h period. In order to understand this KaiABC clockwork, we need to analyze both the macroscopic synchronization of a large number of KaiC hexamers and the microscopic reactions and structural changes in individual KaiC molecules. In the present paper, we explain two coarse-grained theoretical models, the many-molecule (MM) model and the single-molecule (SM) model, to bridge the gap between macroscopic and microscopic understandings. In the simulation results with these models, ATP hydrolysis in the CI domain of KaiC hexamers drives oscillation of individual KaiC hexamers and the ATP hydrolysis is necessary for synchronizing oscillations of a large number of KaiC hexamers. Sensitive temperature dependence of the lifetime of the ADP bound state in the CI domain makes the oscillation period temperature insensitive. ATPase activity is correlated to the frequency of phosphorylation oscillation in the single molecule of KaiC hexamer, which should be the origin of the observed ensemble-level correlation between the ATPase activity and the frequency of phosphorylation oscillation. Thus, the simulation results with the MM and SM models suggest that ATP hydrolysis stochastically occurring in each CI domain of individual KaiC hexamers is a key process for oscillatory behaviors of the ensemble of many KaiC hexamers.
- 著者
- Keisuke Arikawa
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.15, pp.58-74, 2018 (Released:2018-02-27)
- 参考文献数
- 50
- 被引用文献数
- 2
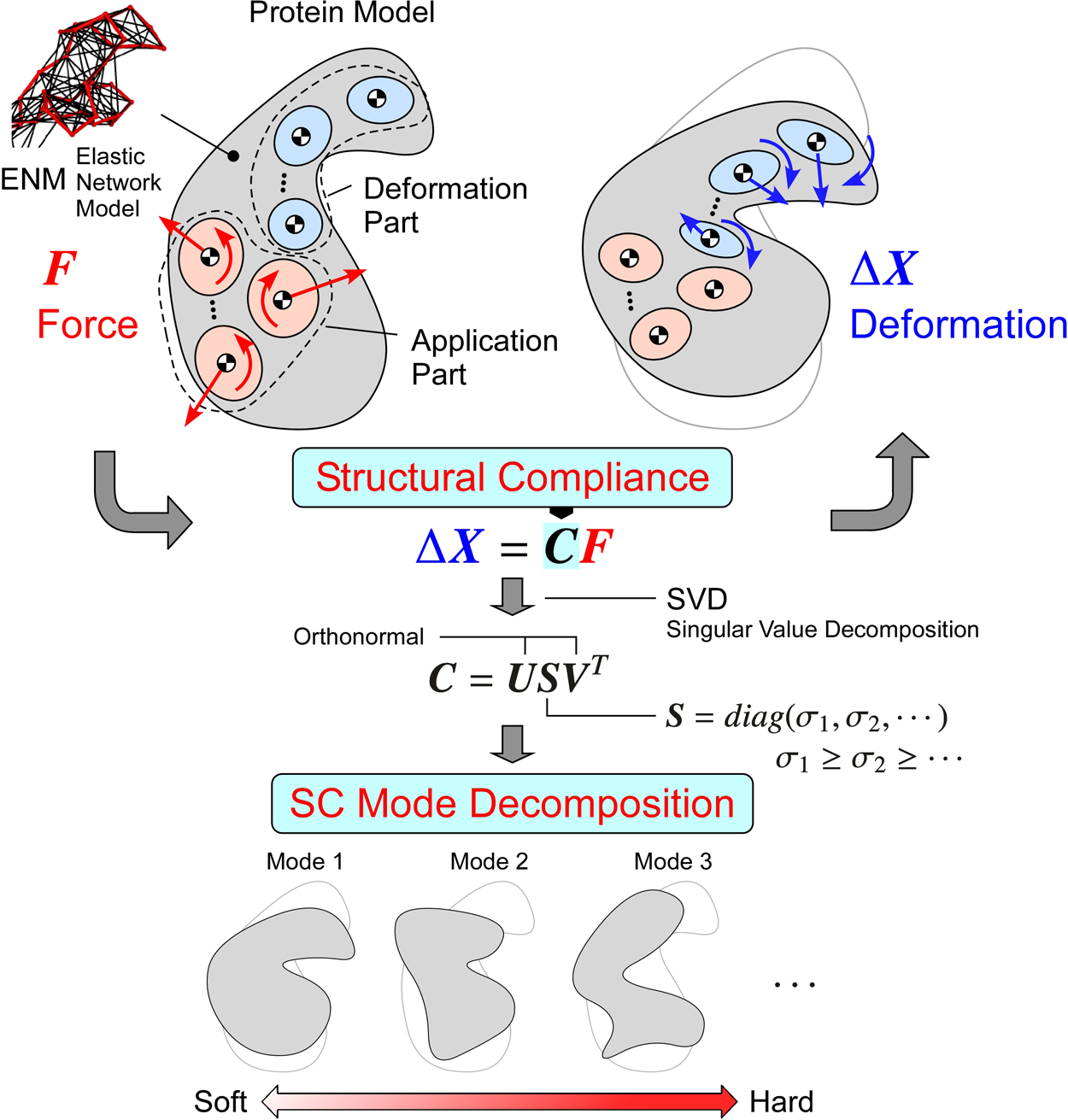
We propose methods for directly analyzing structural compliance (SC) properties of elastic network models of proteins, and we also propose methods for extracting information about motion properties from the SC properties. The analysis of SC properties involves describing the relationships between the applied forces and the deformations. When decomposing the motion according to the magnitude of SC (SC mode decomposition), we can obtain information about the motion properties under the assumption that the lower SC mode motions or the softer motions occur easily. For practical applications, the methods are formulated in a general form. The parts where forces are applied and those where deformations are evaluated are separated from each other for enabling the analyses of allosteric interactions between the specified parts. The parts are specified not only by the points but also by the groups of points (the groups are treated as flexible bodies). In addition, we propose methods for quantitatively evaluating the properties based on the screw theory and the considerations of the algebraic structures of the basic equations expressing the SC properties. These methods enable quantitative discussions about the relationships between the SC mode motions and the motions estimated from two different conformations; they also help identify the key parts that play important roles for the motions by comparing the SC properties with those of partially constrained models. As application examples, lactoferrin and ATCase are analyzed. The results show that we can understand their motion properties through their lower SC mode motions or the softer motions.
- 著者
- Shuya Ishii Madoka Suzuki Shin’ichi Ishiwata Masataka Kawai
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.16, pp.28-40, 2019 (Released:2019-02-02)
- 参考文献数
- 77
- 被引用文献数
- 6
The majority of hypertrophic cardiomyopathy (HCM) is caused by mutations in sarcomere proteins. We examined tropomyosin (Tpm)’s HCM mutants in humans, V95A and D175N, with in vitro motility assay using optical tweezers to evaluate the effects of the Tpm mutations on the actomyosin interaction at the single molecular level. Thin filaments were reconstituted using these Tpm mutants, and their sliding velocity and force were measured at varying Ca2+ concentrations. Our results indicate that the sliding velocity at pCa ≥8.0 was significantly increased in mutants, which is expected to cause a diastolic problem. The velocity that can be activated by Ca2+ decreased significantly in mutants causing a systolic problem. With sliding force, Ca2+ activatable force decreased in V95A and increased in D175N, which may cause a systolic problem. Our results further demonstrate that the duty ratio determined at the steady state of force generation in saturating [Ca2+] decreased in V95A and increased in D175N. The Ca2+ sensitivity and cooperativity were not significantly affected by the mutations. These results suggest that the two mutants modulate molecular processes of the actomyosin interaction differently, but to result in the same pathology known as HCM.
- 著者
- Hiroyuki Terashima Katsumi Imada
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.15, pp.173-178, 2018 (Released:2018-08-22)
- 参考文献数
- 31
- 被引用文献数
- 4
Type III secretion system (T3SS) is a protein translocator complex family including pathogenic injectisome or bacterial flagellum. The inejectisomal T3SS serves to deliver virulence proteins into host cell and the flagellar T3SS constructs the flagellar axial structure. Although earlier studies have provided many findings on the molecular mechanism of the Type III protein export, they were not sufficient to reveal energy transduction mechanism due to difficulties in controlling measurement conditions in vivo. Recently, we developed an in vitro flagellar Type III protein transport assay system using inverted membrane vesicles (IMVs), and analyzed protein export by using the in vitro method. We reproduced protein export of the flagellar T3SS, hook assembly and substrate specificity switch in IMV to a similar extent to what is seen in living cell. Furthermore, we demonstrated that ATP-hydrolysis energy can drive protein transport even in the absence of proton-motive force (PMF). In this mini-review, we will summarize our new in vitro Type III transport assay method and our findings on the molecular mechanism of Type III protein export.
1 0 0 0 OA iMusta4SLC: Database for the structural property and variations of solute carrier transporters
- 著者
- Akiko Higuchi Naoki Nonaka Kei Yura
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.15, pp.94-103, 2018 (Released:2018-04-27)
- 参考文献数
- 50
- 被引用文献数
- 7
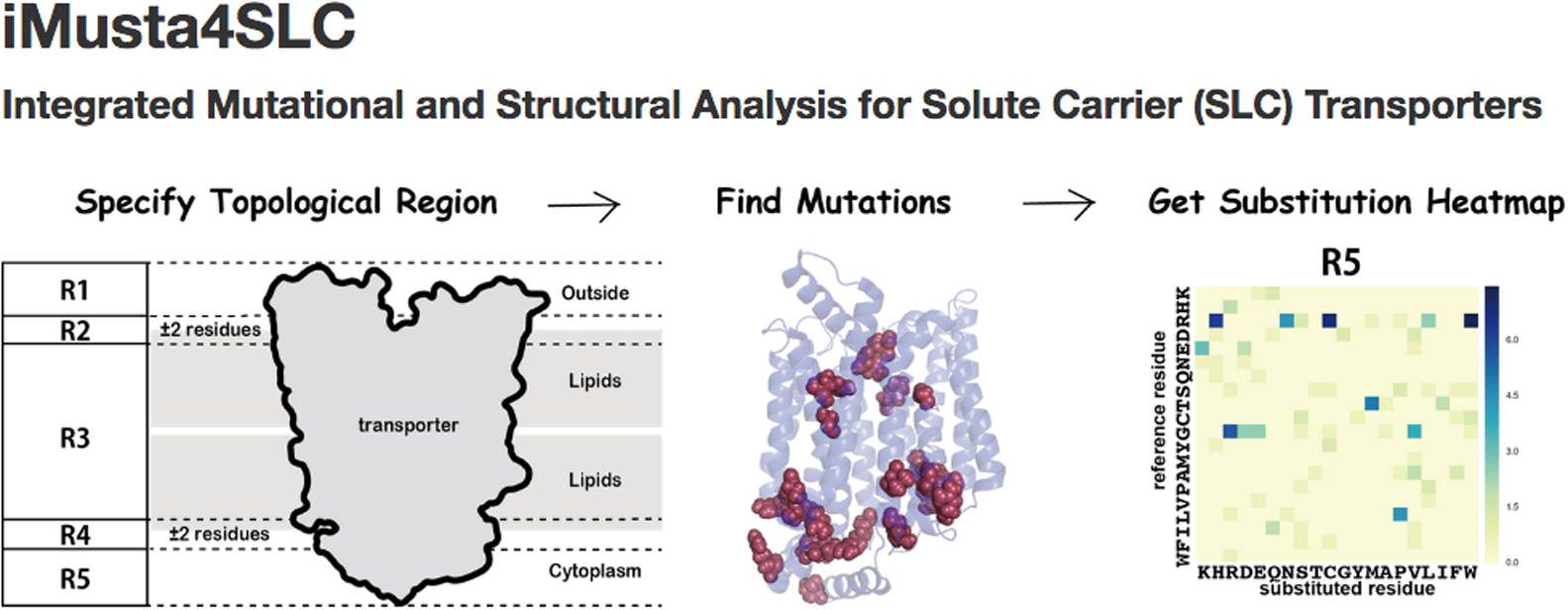
Membrane transporter proteins play important roles in transport of nutrients into the cell, in transport of waste out of the cell, in maintenance of homeostasis, and in signal transduction. Solute carrier (SLC) transporter is the superfamily, which has the largest number of genes (>400 in humans) in membrane transporter and consists of 52 families. SLC transporters carry a wide variety of substrates such as amino acids, peptides, saccharides, ions, neurotransmitters, lipids, hormones and related materials. Despite the apparent importance for the substrate transport, the information of sequence variation and three-dimensional structures have not been integrated to the level of providing new knowledge on the relationship to, for instance, diseases. We, therefore, built a new database named iMusta4SLC, which is available at http://cib.cf.ocha.ac.jp/slc/, that connected the data of structural properties and of pathogenic mutations on human SLC transporters. iMusta4SLC helps to investigate the structural features of pathogenic mutations on SLC transporters. With this database, we found that the mutations at the conserved arginine were frequently involved in diseases, and were located at a border between the membrane and the cytoplasm. Especially in SLC families 2 and 22, the conserved residues formed a large cluster at the border. In SLC2A1, one third of the reported pathogenic missense mutations were found in this conserved cluster.
- 著者
- Jun Tamogami Takashi Kikukawa Toshifumi Nara Makoto Demura Tomomi Kimura-Someya Mikako Shirouzu Shigeyuki Yokoyama Seiji Miyauchi Kazumi Shimono Naoki Kamo
- 出版者
- 一般社団法人 日本生物物理学会
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.14, pp.49-55, 2017 (Released:2017-03-18)
- 参考文献数
- 25
- 被引用文献数
- 7
A spectrally silent change is often observed in the photocycle of microbial rhodopsins. Here, we suggest the presence of two O intermediates in the photocycle of Acetabularia rhodopsin II (ARII or also called Ace2), a light-driven algal proton pump from Acetabularia acetabulum. ARII exhibits a photocycle including a quasi-equilibrium state of M, N, and O (M<=>N<=>O→) at near neutral and above pH values. However, acidification of the medium below pH ~5.5 causes no accumulation of N, resulting in that the photocycle of ARII can be described as an irreversible scheme (M→O→). This may facilitate the investigation of the latter part of the photocycle, especially the rise and decay of O, during which molecular events have not been sufficiently understood. Thus we analyzed the photocycle under acidic conditions (pH ≤ 5.5). Analysis of the absorbance change at 610 nm, which mainly monitors the fractional concentration changes of K and O, was performed and revealed a photocycle scheme containing two sequential O-states with the different molar extinction coefficients. These photoproducts, termed O1 and O2, may be even produced at physiological pH, although they are not clearly observed under this condition due to the existence of a long M-N-O equilibrium.
- 著者
- Yumeka Yamauchi Masae Konno Shota Ito Satoshi P. Tsunoda Keiichi Inoue Hideki Kandori
- 出版者
- 一般社団法人 日本生物物理学会
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.14, pp.57-66, 2017 (Released:2017-05-20)
- 参考文献数
- 58
- 被引用文献数
- 35
Microbial rhodopsins are membrane proteins found widely in archaea, eubacteria and eukaryotes (fungal and algal species). They have various functions, such as light-driven ion pumps, light-gated ion channels, light sensors and light-activated enzymes. A light-driven proton pump bacteriorhodopsin (BR) contains a DTD motif at positions 85, 89, and 96, which is unique to archaeal proton pumps. Recently, channelrhodopsins (ChRs) containing the DTD motif, whose sequential identity is ~20% similar to BR and to cation ChRs in Chlamydomonas reinhardtii (CrCCRs), were found. While extensive studies on ChRs have been performed with CrCCR2, the molecular properties of DTD ChRs remain an intrigue. In this paper, we studied a DTD rhodopsin from G. theta (GtCCR4) using electrophysiological measurements, flash photolysis, and low-temperature difference FTIR spectroscopy. Electrophysiological measurements clearly showed that GtCCR4 functions as a light-gated cation channel, similar to other G. theta DTD ChRs (GtCCR1-3). Light-driven proton pump activity was also suggested for GtCCR4. Both electrophysiological and flash photolysis experiments showed that channel closing occurs upon reprotonation of the Schiff base, suggesting that the dynamics of retinal and channels are tightly coupled in GtCCR4. From Fourier transform infrared (FTIR) spectroscopy at 77 K, we found that the primary reaction is an all-trans to a 13-cis photoisomerization, like other microbial rhodopsins, although perturbations in the secondary structure were much smaller in GtCCR4 than in CrCCR2.
- 著者
- Kei Yura
- 出版者
- 一般社団法人 日本生物物理学会
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.13, pp.245-247, 2016 (Released:2016-11-18)
- 参考文献数
- 13
- 被引用文献数
- 1 2
- 著者
- Oanh T. P. Kim Manh D. Le Hoang X. Trinh Hai V. Nong
- 出版者
- 一般社団法人 日本生物物理学会
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.13, pp.173-180, 2016 (Released:2016-07-14)
- 参考文献数
- 42
- 被引用文献数
- 22
Tumor necrosis factor-alpha (TNF-α) is a cytokine that plays an important role in inflammatory process and tumor development. Recent studies demonstrate that triterpene saponins from Vietnamese ginseng are efficient inhibitors of TNF-α. But the interactions between TNF-α and the saponins are still unclear. In this study, molecular docking and molecular dynamics simulations of TNF-α with three different triterpene saponins (majonoside R2, vina-ginsenoside R1 and vina-ginsenoside R2) were performed to evaluate their binding ability. Our results showed that the triterpene saponins have a good binding affinity with protein TNF-α. The saponins were docked to the pore at the top of the “bell” or “cone” shaped TNF-α trimer and the complexes were structurally stable during 100 ns molecular dynamics simulation. The predicted binding sites would help to subsequently investigate the inhibitory mechanism of triterpene saponins.
- 著者
- Hiroo Imai Nami Suzuki-Hashido Yoshiro Ishimaru Takanobu Sakurai Lijie Yin Wenshi Pan Masaji Ishiguro Katsuyoshi Masuda Keiko Abe Takumi Misaka Hirohisa Hirai
- 出版者
- 一般社団法人 日本生物物理学会
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.13, pp.165-171, 2016 (Released:2016-07-14)
- 参考文献数
- 14
- 被引用文献数
- 1 5
In mammals, bitter taste is mediated by TAS2Rs, which belong to the family of seven transmembrane G protein-coupled receptors. Since TAS2Rs are directly involved in the interaction between mammals and their dietary sources, it is likely that these genes evolved to reflect species-specific diets during mammalian evolution. Here, we analyzed the amino acids responsible for the difference in sensitivities of TAS2R16s of various primates using a cultured cell expression system. We found that the sensitivity of TAS2R16 varied due to several amino acid residues. Mutation of amino acid residues at E86T, L247M, and V260F in human and langur TAS2R16 for mimicking the macaque TAS2R16 decreased the sensitivity of the receptor in an additive manner, which suggests its contribution to the potency of salicin, possibly via direct interaction. However, mutation of amino acid residues 125 and 133 in human TAS2R16, which are situated in helix 4, to the macaque sequence increased the sensitivity of the receptor. These results suggest the possibility that bitter taste sensitivities evolved independently by replacing specific amino acid residues of TAS2Rs in different primate species to adapt to species-specific food.
- 著者
- Kazunori D. Yamada Hafumi Nishi Junichi Nakata Kengo Kinoshita
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.13, pp.157-163, 2016 (Released:2016-07-14)
- 参考文献数
- 41
- 被引用文献数
- 7
Functional sites on proteins play an important role in various molecular interactions and reactions between proteins and other molecules. Thus, mutations in functional sites can severely affect the overall phenotype. Progress of genome sequencing projects has yielded a wealth of information on single nucleotide variants (SNVs), especially those with less than 1% minor allele frequency (rare variants). To understand the functional influence of genetic variants at a protein level, we investigated the relationship between SNVs and protein functional sites in terms of minor allele frequency and the structural position of variants. As a result, we observed that SNVs were less abundant at ligand binding sites, which is consistent with a previous study on SNVs and protein interaction sites. Additionally, we found that non-rare variants tended to be located slightly apart from enzyme active sites. Examination of non-rare variants revealed that most of the mutations resulted in moderate changes of the physico-chemical properties of amino acids, suggesting the existence of functional constraints. In conclusion, this study shows that the mapping of genetic variants on protein structures could be a powerful approach to evaluate the functional impact of rare genetic variations.
- 著者
- Masayuki Oda Takeshi Tsumuraya Ikuo Fujii
- 出版者
- 一般社団法人 日本生物物理学会
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.13, pp.135-138, 2016 (Released:2016-07-14)
- 参考文献数
- 14
- 被引用文献数
- 2
We analyzed the correlation between the conformational strain and the binding kinetics in antigen-antibody interactions. The catalytic antibodies 6D9, 9C10, and 7C8 catalyze the hydrolysis of a nonbioactive chloramphenicol monoester derivative to generate a bioactive chloramphenicol. The crystal structure of 6D9 complexed with a transition-state analog (TSA) suggests that 6D9 binds the substrate to change the conformation of the ester moiety to a thermodynamically unstable twisted conformation, enabling the substrate to reach the transition state during catalysis. The present binding kinetic analysis showed that the association rate for 6D9 binding to the substrate was much lower than that to TSA, whereas those for 9C10 and 7C8 binding were similar to those to TSA. Considering that 7C8 binds to the substrate with little conformational change in the substrate, the slow association rate observed in 6D9 could be attributed to the conformational strain in the substrate.
- 著者
- Yuka Suzuki Kei Yura
- 出版者
- 一般社団法人 日本生物物理学会
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.13, pp.127-134, 2016 (Released:2016-07-14)
- 参考文献数
- 26
- 被引用文献数
- 4
We investigated the effect of ATP binding to GroEL and elucidated a role of ATP in the conformational change of GroEL. GroEL is a tetradecamer chaperonin that helps protein folding by undergoing a conformational change from a closed state to an open state. This conformational change requires ATP, but does not require the hydrolysis of the ATP. The following three types of conformations are crystalized and the atomic coordinates are available; closed state without ATP, closed state with ATP and open state with ADP. We conducted simulations of the conformational change using Elastic Network Model from the closed state without ATP targeting at the open state, and from the closed state with ATP targeting at the open state. The simulations emphasizing the lowest normal mode showed that the one started with the closed state with ATP, rather than the one without ATP, reached a conformation closer to the open state. This difference was mainly caused by the changes in the positions of residues in the initial structure rather than the changes in “connectivity” of residues within the subunit. Our results suggest that ATP should behave as an insulator to induce conformation population shift in the closed state to the conformation that has a pathway leading to the open state.
- 著者
- Georgios Iakovou Stephen Laycock Steven Hayward
- 出版者
- 一般社団法人 日本生物物理学会
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.13, pp.97-103, 2016 (Released:2016-07-14)
- 参考文献数
- 37
- 被引用文献数
- 5
Interactive haptics-assisted docking provides a virtual environment for the study of molecular complex formation. It enables the user to interact with the virtual molecules, experience the interaction forces via their sense of touch, and gain insights about the docking process itself. Here we use a recently developed haptics software tool, Haptimol_RD, for the rigid docking of protein subunits to form complexes. Dimers, both homo and hetero, are loaded into the software with their subunits separated in space for the purpose of assessing whether they can be brought back into the correct docking pose via rigid-body movements. Four dimers were classified into two types: two with an interwinding subunit interface and two with a non-interwinding subunit interface. It was found that the two with an interwinding interface could not be docked whereas the two with the non-interwinding interface could be. For the two that could be docked a “sucking” effect could be felt on the haptic device when the correct binding pose was approached which is associated with a minimum in the interaction energy. It is clear that for those that could not be docked, the conformation of one or both of the subunits must change upon docking. This leads to the steric-based concept of a locked or non-locked interface. Non-locked interfaces have shapes that allow the subunits to come together or come apart without the necessity of intra-subunit conformational change, whereas locked interfaces require a conformational change within one or both subunits for them to be able to come apart.
- 著者
- Kentaro Ishii Masanori Noda Susumu Uchiyama
- 出版者
- 一般社団法人 日本生物物理学会
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.13, pp.87-95, 2016 (Released:2016-07-14)
- 参考文献数
- 32
- 被引用文献数
- 32
The interactions of small molecules with proteins (protein–ligand interactions) mediate various biological phenomena including signal transduction and protein transcription and translation. Synthetic compounds such as drugs can also bind to target proteins, leading to the inhibition of protein–ligand interactions. These interactions typically accompany association–dissociation equilibrium according to the free energy difference between free and bound states; therefore, the quantitative biophysical analysis of the interactions, which uncovers the stoichiometry and dissociation constant, is important for understanding biological reactions as well as for rational drug development. Mass spectrometry (MS) has been used to determine the precise molecular masses of molecules. Recent advancements in MS enable us to determine the molecular masses of protein–ligand complexes without disrupting the non-covalent interactions through the gentle desolvation of the complexes by increasing the vacuum pressure of a chamber in a mass spectrometer. This method is called MS under non-denaturing conditions or native MS and allows the unambiguous determination of protein–ligand interactions. Under a few assumptions, MS has also been applied to determine the dissociation constants for protein–ligand interactions. The structural information of a protein–ligand interaction, such as the location of the interaction and conformational change in a protein, can also be analyzed using hydrogen/deuterium exchange MS. In this paper, we briefly describe the history, principle, and recent applications of MS for the study of protein–ligand interactions.