- 著者
- María del Carmen Marín Alexander L. Jaffe Patrick T. West Masae Konno Jillian F. Banfield Keiichi Inoue
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.e201023, (Released:2023-03-08)
- 被引用文献数
- 1
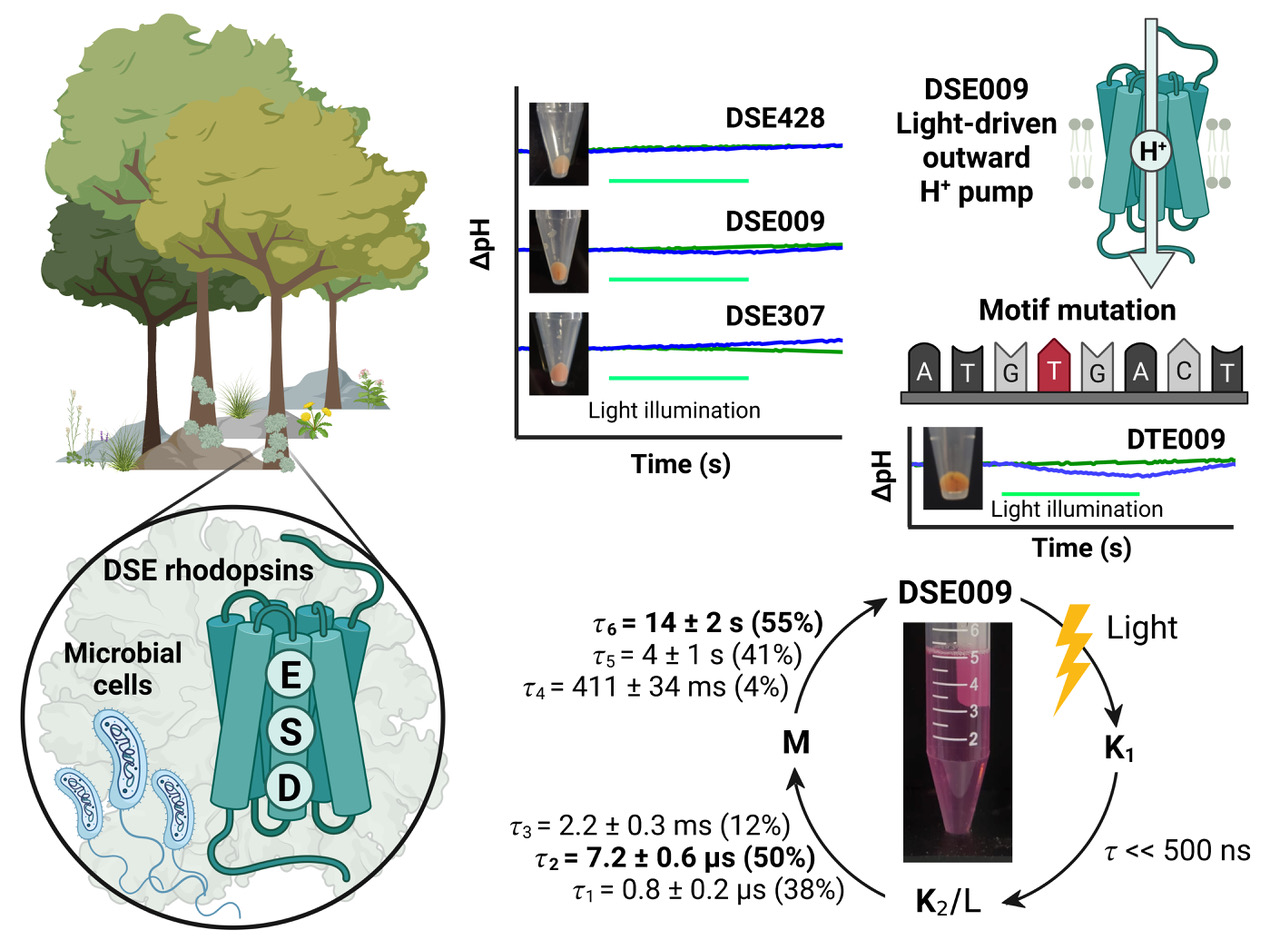
Microbial rhodopsins are photoreceptive transmembrane proteins that transport ions or regulate other intracellular biological processes. Recent genomic and metagenomic analyses found many microbial rhodopsins with unique sequences distinct from known ones. Functional characterization of these new types of microbial rhodopsins is expected to expand our understanding of their physiological roles. Here, we found microbial rhodopsins having a DSE motif in the third transmembrane helix from members of the Actinobacteria. Although the expressed proteins exhibited blue–green light absorption, either no or extremely small outward H+ pump activity was observed. The turnover rate of the photocycle reaction of the purified proteins was extremely slow compared to typical H+ pumps, suggesting these rhodopsins would work as photosensors or H+ pumps whose activities are enhanced by an unknown regulatory system in the hosts. The discovery of this rhodopsin group with the unique motif and functionality expands our understanding of the biological role of microbial rhodopsins.
- 著者
- Editorial team for the Special Issue on Oosawa’s Lectures
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.18, no.Supplemental, pp.S001-S002, 2021 (Released:2021-12-10)
- 参考文献数
- 3
- 被引用文献数
- 1
- 著者
- Daniel Dai Muneyoshi Ichikawa Katya Peri Reid Rebinsky Khanh Huy Bui
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.17, pp.71-85, 2020 (Released:2020-07-22)
- 参考文献数
- 48
- 被引用文献数
- 13 28
Cilia or flagella of eukaryotes are small micro-hair like structures that are indispensable to single-cell motility and play an important role in mammalian biological processes. Cilia or flagella are composed of nine doublet microtubules surrounding a pair of singlet microtubules called the central pair (CP). Together, this arrangement forms a canonical and highly conserved 9+2 axonemal structure. The CP, which is a unique structure exclusive to motile cilia, is a pair of structurally dimorphic singlet microtubules decorated with numerous associated proteins. Mutations of CP-associated proteins cause several different physical symptoms termed as ciliopathies. Thus, it is crucial to understand the architecture of the CP. However, the protein composition of the CP was poorly understood. This was because the traditional method of identification of CP proteins was mostly limited by available Chlamydomonas mutants of CP proteins. Recently, more CP protein candidates were presented based on mass spectrometry results, but most of these proteins were not validated. In this study, we re-evaluated the CP proteins by conducting a similar comprehensive CP proteome analysis comparing the mass spectrometry results of the axoneme sample prepared from Chlamydomonas strains with and without CP complex. We identified a similar set of CP protein candidates and additional new 11 CP protein candidates. Furthermore, by using Chlamydomonas strains lacking specific CP sub-structures, we present a more complete model of localization for these CP proteins. This work has established a new foundation for understanding the function of the CP complex in future studies.
- 著者
- Tony Z. Jia Kuhan Chandru
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.20, no.1, pp.e200012, 2023 (Released:2023-03-04)
- 参考文献数
- 92
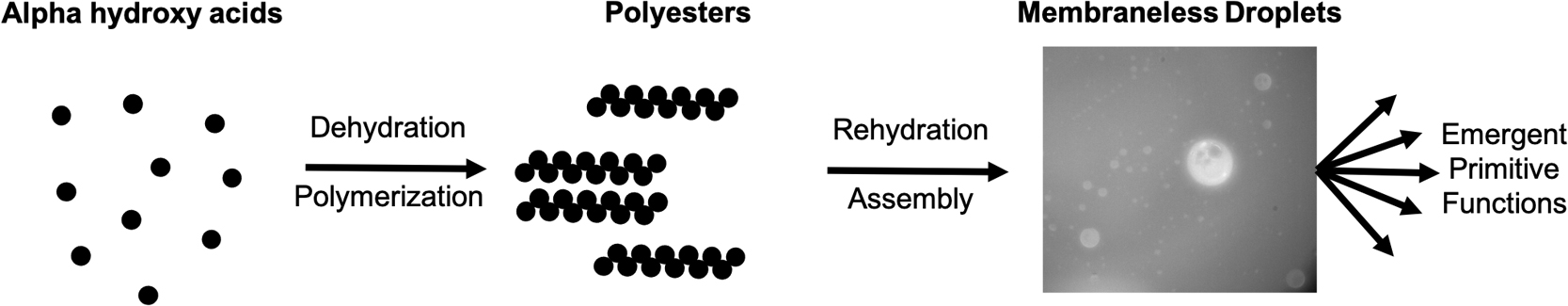
While it is often believed that the origins of life required participation of early biomolecules, it has been recently proposed that “non-biomolecules”, which would have been just as, if not more, abundant on early Earth, could have played a part. In particular, recent research has highlighted the various ways by which polyesters, which do not participate in modern biology, could have played a major role during the origins of life. Polyesters could have been synthesized readily on early Earth through simple dehydration reactions at mild temperatures involving abundant “non-biological” alpha hydroxy acid (AHA) monomers. This dehydration synthesis process results in a polyester gel, which upon further rehydration, can assemble into membraneless droplets proposed to be protocell models. These proposed protocells can provide functions to a primitive chemical system, such as analyte segregation or protection, which could have further led to chemical evolution from prebiotic chemistry to nascent biochemistry. Here, to further shed light into the importance of “non-biomolecular” polyesters at the origins of life and to highlight future directions of study, we review recent studies which focus on primitive synthesis of polyesters from AHAs and assembly of these polyesters into membraneless droplets. Specifically, most of the recent progress in this field in the last five years has been led by laboratories in Japan, and these will be especially highlighted. This article is based on an invited presentation at the 60th Annual Meeting of the Biophysical Society of Japan held in September, 2022 as an 18th Early Career Awardee.
- 著者
- Izuru Kawamura Hayato Seki Seiya Tajima Yoshiteru Makino Arisu Shigeta Takashi Okitsu Akimori Wada Akira Naito Yuki Sudo
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.18, pp.177-185, 2021 (Released:2021-08-07)
- 参考文献数
- 53
- 被引用文献数
- 6
Middle rhodopsin (MR) found from the archaeon Haloquadratum walsbyi is evolutionarily located between two different types of rhodopsins, bacteriorhodopsin (BR) and sensory rhodopsin II (SRII). Some isomers of the chromophore retinal and the photochemical reaction of MR are markedly different from those of BR and SRII. In this study, to obtain the structural information regarding its active center (i.e., retinal), we subjected MR embedded in lipid bilayers to solid-state magic-angle spinning nuclear magnetic resonance (NMR) spectroscopy. The analysis of the isotropic 13C chemical shifts of the retinal chromophore revealed the presence of three types of retinal configurations of dark-adapted MR: (13-trans, 15-anti (all-trans)), (13-cis, 15-syn), and 11-cis isomers. The higher field resonance of the 20-C methyl carbon in the all-trans retinal suggested that Trp182 in MR has an orientation that is different from that in other microbial rhodopsins, owing to the changes in steric hindrance associated with the 20-C methyl group in retinal. 13Cζ signals of Tyr185 in MR for all-trans and 13-cis, 15-syn isomers were discretely observed, representing the difference in the hydrogen bond strength of Tyr185. Further, 15N NMR analysis of the protonated Schiff base corresponding to the all-trans and 13-cis, 15-syn isomers in MR showed a strong electrostatic interaction with the counter ion. Therefore, the resulting structural information exhibited the property of stable retinal conformations of dark-adapted MR.
- 著者
- Kumari Sushmita Sunita Sharma Manish Singh Kaushik Suneel Kateriya
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.e201008, (Released:2023-01-24)
- 被引用文献数
- 2
Rhodopsins have been extensively employed for optogenetic regulation of bioelectrical activity of excitable cells and other cellular processes across biological systems. Various strategies have been adopted to attune the cellular processes at the desired subcellular compartment (plasma membrane, endoplasmic reticulum, Golgi, mitochondria, lysosome) within the cell. These strategies include- adding signal sequences, tethering peptides, specific interaction sites, or mRNA elements at different sites in the optogenetic proteins for plasma membrane integration and subcellular targeting. However, a single approach for organelle optogenetics was not suitable for the relevant optogenetic proteins and often led to the poor expression, mislocalization, or altered physical and functional properties. Therefore, the current study is focused on the native subcellular targeting machinery of algal rhodopsins. The N- and C-terminus signal prediction led to the identification of rhodopsins with diverse organelle targeting signal sequences for the nucleus, mitochondria, lysosome, endosome, vacuole, and cilia. Several identified channelrhodopsins and ion-pumping rhodopsins possess effector domains associated with DNA metabolism (repair, replication, and recombination) and gene regulation. The identified algal rhodopsins with diverse effector domains and encoded native subcellular targeting sequences hold immense potential to establish expanded organelle optogenetic regulation and associated cellular signaling.
- 著者
- Damien Hall Adam S. Foster
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.19, pp.e190016, 2022 (Released:2022-05-18)
- 参考文献数
- 97
- 被引用文献数
- 9
High speed atomic force microscopy (HS-AFM) is, in principle, capable of yielding nanometer level detail about the surface of static structures. However, for highly dynamic samples HS-AFM may struggle with the correct feature assignment both within and between frames. Feature assignment in HS-AFM is dependent on (i) the intrinsic sampling rate, and (ii) the rate of internal redistribution of the sample. Whilst the first quantity (the sampling rate) is defined by the device parameters, the second quantity is frequently unknown, and is often the desired target of the measurement. This work examines how, even in the absence of gross cell morphological change, the rapid dynamics of living cell membranes, may impose an upper spatial limit to the frame-to-frame assignment of cell micro-topography and other related properties (such as local elasticity) whose motion may be described stochastically. Such a practical maximum may prove useful in the setup of HS-AFM experiments involving dynamic surfaces thereby facilitating selection of the most parsimonious relationship between observation size, image pixilation and sampling rates. To assist with performing the described calculations a graphical user interface-based software package called HS-AFM UGOKU is made freely available.
- 著者
- Takeharu Nagai Shunsuke Chikuma Kenjiro Hanaoka
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.BSJ-2020018, (Released:2020-08-28)
- 著者
- Tetsuichi Wazawa Takeharu Nagai
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.20, no.3, pp.e200030, 2023 (Released:2023-07-12)
- 参考文献数
- 43
Ion currents associated with channel proteins in the presence of membrane potential are ubiquitous in cellular and organelle membranes. When an ion current occurs through a channel protein, Joule heating should occur. However, this Joule heating seems to have been largely overlooked in biology. Here we show theoretical investigation of Joule heating involving channel proteins in biological processes. We used electrochemical potential to derive the Joule’s law for an ion current through an ion transport protein in the presence of membrane potential, and we suggest that heat production and absorption can occur. Simulation of temperature distribution around a single channel protein with the Joule heating revealed that the temperature increase was as small as <10–3 K, although an ensemble of channel proteins was suggested to exhibit a noticeable temperature increase. Thereby, we theoretically investigated the Joule heating of systems containing ensembles of channel proteins. Nerve is known to undergo rapid heat production followed by heat absorption during the action potential, and our simulation of Joule heating for a squid giant axon combined with the Hodgkin-Huxley model successfully reproduced the feature of the heat. Furthermore, we extended the theory of Joule heating to uncoupling protein 1 (UCP1), a solute carrier family transporter, which is important to the non-shivering thermogenesis in brown adipose tissue mitochondria (BATM). Our calculations showed that the Joule heat involving UCP1 was comparable to the literature calorimetry data of BATM. Joule heating of ion transport proteins is likely to be one of important mechanisms of cellular thermogenesis.
- 著者
- Emi Hibino Masaru Hoshino
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.17, pp.86-93, 2020 (Released:2020-08-20)
- 参考文献数
- 34
- 被引用文献数
- 6 8
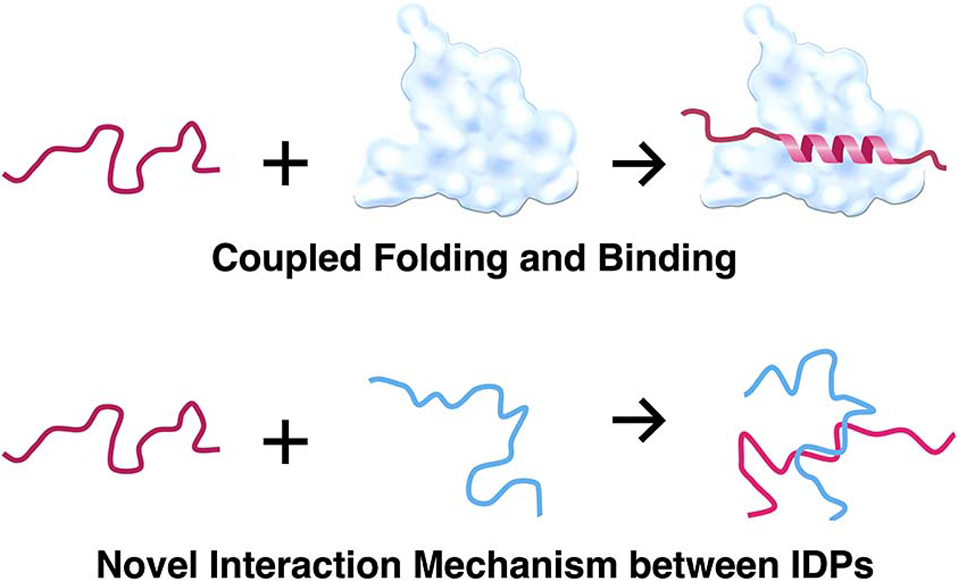
An increasing number of proteins, which have neither regular secondary nor well-defined tertiary structures, have been found to be present in cells. The structure of these proteins is highly flexible and disordered under physiological (native) conditions, and they are called “intrinsically disordered” proteins (IDPs). Many of the IDPs are involved in interactions with other biomolecules such as DNA, RNA, carbohydrates, and proteins. While these IDPs are largely unstructured by themselves, marked conformational changes often occur upon binding to an interacting partner, which is known as the “coupled folding and binding mechanism”, which enable them to change the conformation to become compatible with the shape of the multiple target biomolecules. We have studied the structure and interaction of eukaryotic transcription factors Sp1 and TAF4, and found that both of them have long intrinsically disordered regions (IDRs). One of the IDRs in Sp1 exhibited homo-oligomer formation. In addition, the same region was used for the interaction with another IDR found in the TAF4 molecule. In both cases, we have not detected any significant conformational change in that region, suggesting a prominent and novel binding mode for IDPs/IDRs, which are not categorized by the well-accepted concept of the coupled folding and binding mechanism.
- 著者
- Katsumasa Irie
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.18, pp.274-283, 2021 (Released:2021-12-04)
- 参考文献数
- 38
- 被引用文献数
- 4
Prokaryotic channels play an important role in the structural biology of ion channels. At the end of the 20th century, the first structure of a prokaryotic ion channel was revealed. Subsequently, the reporting of structures of various prokaryotic ion channels have provided fundamental insights into the structure of ion channels of higher organisms. Voltage-dependent Ca2+ channels (Cavs) are indispensable for coupling action potentials with Ca2+ signaling. Similar to other proteins, Cavs were predicted to have a prokaryotic counterpart; however, it has taken more than 20 years for one to be identified. The homotetrameric channel obtained from Meiothermus ruber generates the calcium ion specific current, so it is named as CavMr. Its selectivity filter contains a smaller number of negatively charged residues than mutant Cavs generated from other prokaryotic channels. CavMr belonged to a different cluster of phylogenetic trees than canonical prokaryotic cation channels. The glycine residue of the CavMr selectivity filter is a determinant for calcium selectivity. This glycine residue is conserved among eukaryotic Cavs, suggesting that there is a universal mechanism for calcium selectivity. A family of homotetrameric channels has also been identified from eukaryotic unicellular algae, and the investigation of these channels can help to understand the mechanism for ion selection that is conserved from prokaryotes to eukaryotes.
- 著者
- Toshiki Yagi Akiyuki Toda Muneyoshi Ichikawa Genji Kurisu
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.20, no.1, pp.e200008, 2023 (Released:2023-03-04)
- 参考文献数
- 43
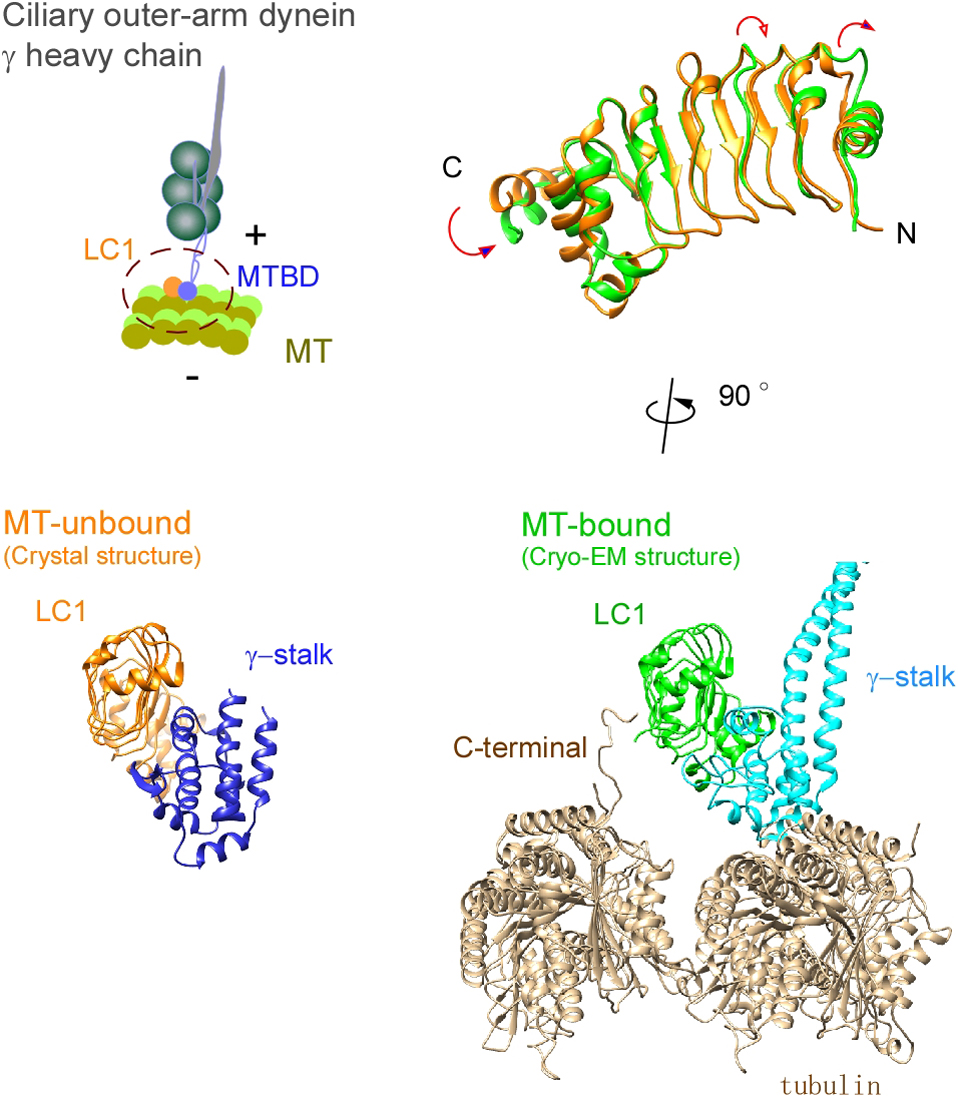
Ciliary bending movements are powered by motor protein axonemal dyneins. They are largely classified into two groups, inner-arm dynein and outer-arm dynein. Outer-arm dynein, which is important for the elevation of ciliary beat frequency, has three heavy chains (α, β, and γ), two intermediate chains, and more than 10 light chains in green algae, Chlamydomonas. Most of intermediate chains and light chains bind to the tail regions of heavy chains. In contrast, the light chain LC1 was found to bind to the ATP-dependent microtubule-binding domain of outer-arm dynein γ-heavy chain. Interestingly, LC1 was also found to interact with microtubules directly, but it reduces the affinity of the microtubule-binding domain of γ-heavy chain for microtubules, suggesting the possibility that LC1 may control ciliary movement by regulating the affinity of outer-arm dyneins for microtubules. This hypothesis is supported by the LC1 mutant studies in Chlamydomonas and Planaria showing that ciliary movements in LC1 mutants were disordered with low coordination of beating and low beat frequency. To understand the molecular mechanism of the regulation of outer-arm dynein motor activity by LC1, X-ray crystallography and cryo-electron microscopy have been used to determine the structure of the light chain bound to the microtubule-binding domain of γ-heavy chain. In this review article, we show the recent progress of structural studies of LC1, and suggest the regulatory role of LC1 in the motor activity of outer-arm dyneins. This review article is an extended version of the Japanese article, The Complex of Outer-arm Dynein Light Chain-1 and the Microtubule-binding Domain of the Heavy Chain Shows How Axonemal Dynein Tunes Ciliary Beating, published in SEIBUTSU BUTSURI Vol. 61, p. 20–22 (2021).
4 0 0 0 OA Identification and structural basis of an enzyme that degrades oligosaccharides in caramel
- 著者
- Toma Kashima Akihiro Ishiwata Kiyotaka Fujita Shinya Fushinobu
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.e200017, (Released:2023-03-29)
- 被引用文献数
- 1
Cooking with fire produces foods containing carbohydrates that are not naturally occurring, such as α-d-fructofuranoside found in caramel. Each of the hundreds of compounds produced by caramelization reactions is considered to possess its own characteristics. Various studies from the viewpoints of biology and biochemistry have been conducted to elucidate some of the scientific characteristics. Here, we review the composition of caramelized sugars and then describe the enzymatic studies that have been conducted and the physiological functions of the caramelized sugar components that have been elucidated. In particular, we recently identified a glycoside hydrolase (GH), GH172 difructose dianhydride I synthase/hydrolase (αFFase1), from oral and intestinal bacteria, which is implicated in the degradation of oligosaccharides in caramel. The structural basis of αFFase1 and its ligands provided many insights. This discovery opened the door to several research fields, including the structural and phylogenetic relationship between the GH172 family enzymes and viral capsid proteins and the degradation of cell membrane glycans of acid-fast bacteria by some αFFase1 homologs. This review article is an extended version of the Japanese article, Identification and Structural Basis of an Enzyme Degrading Oligosaccharides in Caramel, published in SEIBUTSU BUTSURI Vol. 62, p. 184-186 (2022).
4 0 0 0 OA Potassium-selective channelrhodopsins
- 著者
- Elena G. Govorunova Oleg A. Sineshchekov John L. Spudich
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.e201011, (Released:2023-02-04)
- 被引用文献数
- 1
Since their discovery 21 years ago, channelrhodopsins have come of age and have become indispensable tools for optogenetic control of excitable cells such as neurons and myocytes. Potential therapeutic utility of channelrhodopsins has been proven by partial vision restoration in a human patient. Previously known channelrhodopsins are either proton channels, non-selective cation channels almost equally permeable to Na+ and K+ besides protons, or anion channels. Two years ago, we discovered a group of channelrhodopsins that exhibit over an order of magnitude higher selectivity for K+ than for Na+. These proteins, known as “kalium channelrhodopsins” or KCRs, lack the canonical tetrameric selectivity filter found in voltage- and ligand-gated K+ channels, and use a unique selectivity mechanism intrinsic to their individual protomers. Mutant analysis has revealed that the key residues responsible for K+ selectivity in KCRs are located at both ends of the putative cation conduction pathway, and their role has been confirmed by high-resolution KCR structures. Expression of KCRs in mouse neurons and human cardiomyocytes enabled optical inhibition of these cells’ electrical activity. In this minireview we briefly discuss major results of KCR research obtained during the last two years and suggest some directions of future research.
- 著者
- Satoru Tokutomi Satoshi P. Tsunoda
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.19, pp.e190004_1-e190004_3, 2022 (Released:2022-02-26)
- 参考文献数
- 12
- 著者
- Ryohei Kondo Kota Kasahara Takuya Takahashi
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.e190002, (Released:2022-02-08)
- 著者
- Kumiko Hayashi Shinsuke Niwa
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.bppb-v18.026, (Released:2021-10-06)
- 被引用文献数
- 2
- 著者
- Daniel Dai Muneyoshi Ichikawa Katya Peri Reid Rebinsky Khanh Huy Bui
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.BSJ-2019048, (Released:2020-06-16)
- 被引用文献数
- 28
- 著者
- Yudai Yamaoki Takashi Nagata Tomoki Sakamoto Masato Katahira
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.BSJ-2020006, (Released:2020-05-26)
- 被引用文献数
- 6
- 著者
- Shumpei Matsuno Masahito Ohue Yutaka Akiyama
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.17, pp.2-13, 2020 (Released:2020-02-07)
- 参考文献数
- 30
- 被引用文献数
- 3
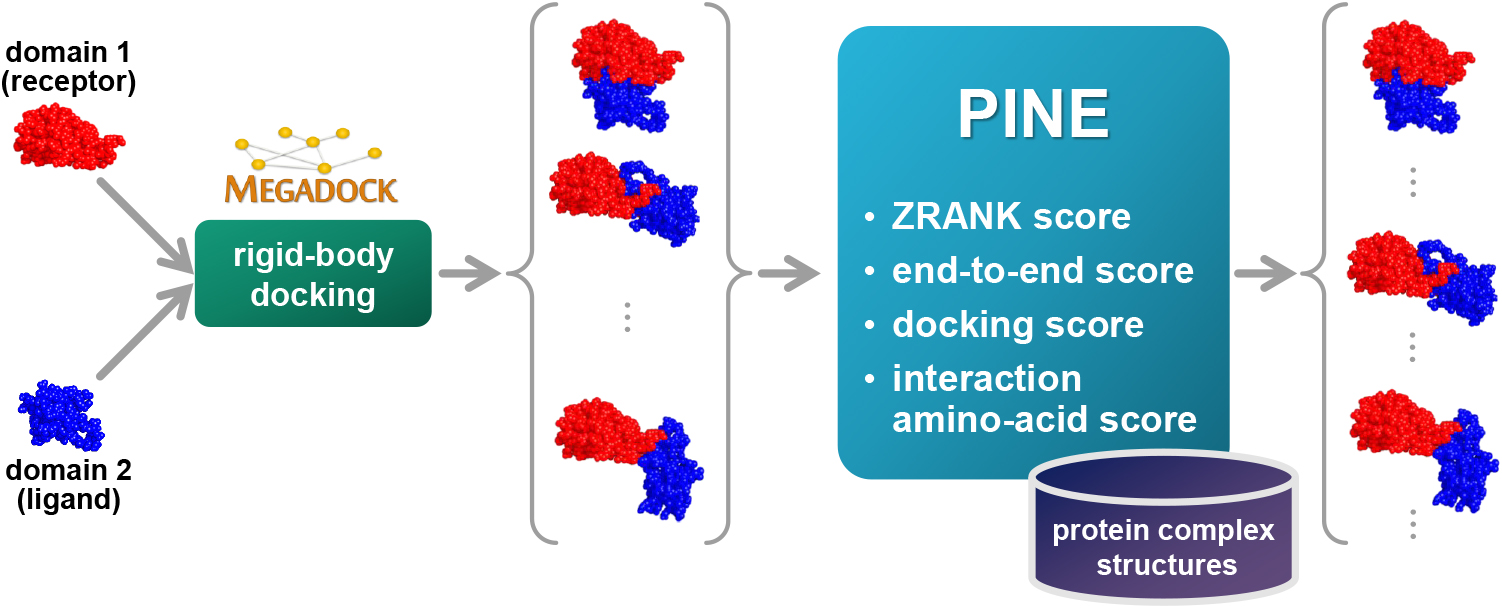
Protein functions can be predicted based on their three-dimensional structures. However, many multidomain proteins have unstable structures, making it difficult to determine the whole structure in biological experiments. Additionally, multidomain proteins are often decomposed and identified based on their domains, with the structure of each domain often found in public databases. Recent studies have advanced structure prediction methods of multidomain proteins through computational analysis. In existing methods, proteins that serve as templates are used for three-dimensional structure prediction. However, when no protein template is available, the accuracy of the prediction is decreased. This study was conducted to predict the structures of multidomain proteins without the need for whole structure templates.We improved structure prediction methods by performing rigid-body docking from the structure of each domain and reranking a structure closer to the correct structure to have a higher value. In the proposed method, the score for the domain-domain interaction obtained without a structural template of the multidomain protein and score for the three-dimensional structure obtained during docking calculation were newly incorporated into the score function. We successfully predicted the structures of 50 of 55 multidomain proteins examined in the test dataset.Interaction residue pair information of the protein-protein complex interface contributes to domain reorganizations even when a structural template for a multidomain protein cannot be obtained. This approach may be useful for predicting the structures of multidomain proteins with important biochemical functions.