- 著者
- Damien Simon Atsushi Mukaiyama Yoshihiko Furuike Shuji Akiyama
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.e190008, (Released:2022-03-30)
- 被引用文献数
- 5
- 著者
- Keisuke Inoue Shoji Takada Tsuyoshi Terakawa
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.19, pp.e190015, 2022 (Released:2022-05-11)
- 参考文献数
- 43
- 被引用文献数
- 2
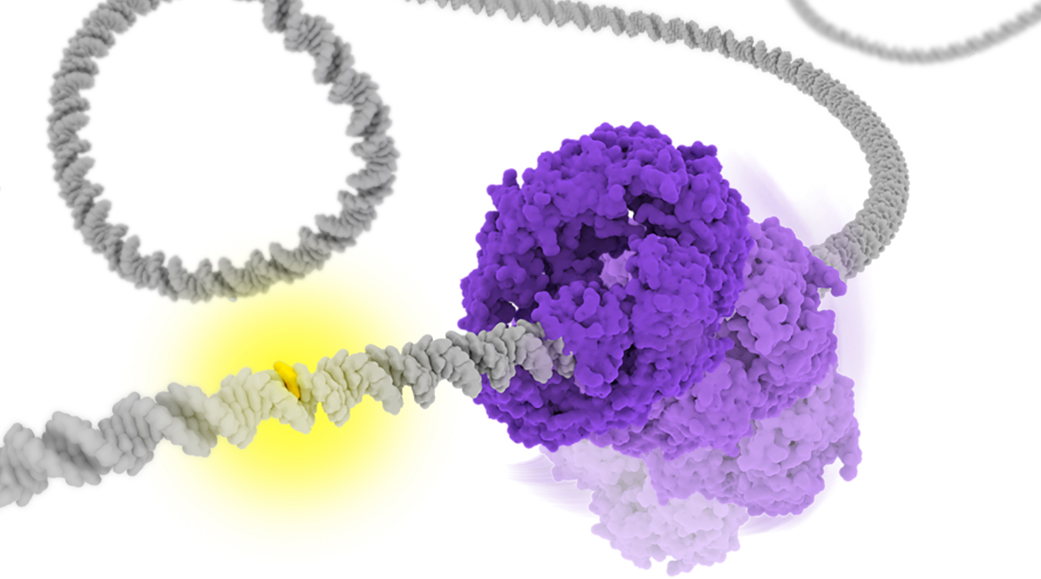
DNA mismatches are frequently generated by various intrinsic and extrinsic factors including DNA replication errors, oxygen species, ultraviolet, and ionizing radiation. These mismatches should be corrected by the mismatches repair (MMR) pathway to maintain genome integrity. In the Escherichia coli (E. coli) MMR pathway, MutS searches and recognizes a base-pair mismatch from millions of base-pairs. Once recognized, ADP bound to MutS is exchanged with ATP, which induces a conformational change in MutS. Previous single-molecule fluorescence microscopy studies have suggested that ADP-bound MutS temporarily slides along double-stranded DNA in a rotation-coupled manner to search a base-pair mismatch and so does ATP-bound MutS in a rotation-uncoupled manner. However, the detailed structural dynamics of the sliding remains unclear. In this study, we performed coarse-grained molecular dynamics simulations of the E. coli MutS bound on DNA in three different conformations: ADP-bound (), ATP-bound open clamp (), and ATP-bound closed clamp () conformations. In the simulations, we observed conformation-dependent diffusion of MutS along DNA. and diffused along DNA in a rotation-coupled manner with rare and frequent groove-crossing events, respectively. In the groove-crossing events, MutS overcame an edge of a groove and temporarily diffused in a rotation-uncoupled manner. It was also indicated that mismatch searches by is inefficient in terms of mismatch checking even though it diffuses along DNA and reaches unchecked regions more rapidly than .
2 0 0 0 OA Beyond multi-disciplinary and cross-scale analyses of the cyanobacterial circadian clock system
- 著者
- Shuji Akiyama Hironari Kamikubo
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.18, pp.267-268, 2021 (Released:2021-11-27)
- 参考文献数
- 10
2 0 0 0 OA Limitations of the ABEGO-based backbone design: ambiguity between αα-corner and αα-hairpin
- 著者
- Koya Sakuma
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.bppb-v18.017, (Released:2021-05-28)
- 著者
- Yutaka Maruyama Hiroshi Takano Ayori Mitsutake
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.16, pp.407-429, 2019 (Released:2019-11-29)
- 参考文献数
- 182
- 被引用文献数
- 6
Molecular dynamics simulation is a fruitful tool for investigating the structural stability, dynamics, and functions of biopolymers at an atomic level. In recent years, simulations can be performed on time scales of the order of milliseconds using specialpurpose systems. Since the most stable structure, as well as meta-stable structures and intermediate structures, is included in trajectories in long simulations, it is necessary to develop analysis methods for extracting them from trajectories of simulations. For these structures, methods for evaluating the stabilities, including the solvent effect, are also needed. We have developed relaxation mode analysis to investigate dynamics and kinetics of simulations based on statistical mechanics. We have also applied the three-dimensional reference interaction site model theory to investigate stabilities with solvent effects. In this paper, we review the results for designing amino-acid substitution of the 10-residue peptide, chignolin, to stabilize the misfolded structure using these developed analysis methods.
- 著者
- Teppei Sugimoto Kota Katayama Hideki Kandori
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.bppb-v18.012, (Released:2021-04-16)
- 被引用文献数
- 6
- 著者
- Akira R. Kinjo
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.12, pp.117-119, 2015 (Released:2015-12-11)
- 参考文献数
- 16
- 被引用文献数
- 1 1
The direct-coupling analysis is a powerful method for protein contact prediction, and enables us to extract “direct” correlations between distant sites that are latent in “indirect” correlations observed in a protein multiple-sequence alignment. I show that the direct correlation can be obtained by using a formulation analogous to the Ornstein-Zernike integral equation in liquid theory. This formulation intuitively illustrates how the indirect or apparent correlation arises from an infinite series of direct correlations, and provides interesting insights into protein structure prediction.
- 著者
- Yujiro Nagasaka Shoko Hososhima Naoko Kubo Takashi Nagata Hideki Kandori Keiichi Inoue Hiromu Yawo
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.BSJ-2020007, (Released:2020-06-09)
- 被引用文献数
- 5
2 0 0 0 OA Consistency principle for protein design
- 著者
- Rie Koga Nobuyasu Koga
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.16, pp.304-309, 2019 (Released:2019-11-29)
- 参考文献数
- 38
- 被引用文献数
- 7
Protein design holds promise for applications such as the control of cells, therapeutics, new enzymes and protein-based materials. Recently, there has been progress in rational design of protein molecules, and a lot of attempts have been made to create proteins with functions of our interests. The key to the progress is the development of methods for controlling desired protein tertiary structures with atomic-level accuracy. A theory for protein folding, the consistency principle, proposed by Nobuhiro Go in 1983, was a compass for the development. Anfinsen hypothesized that proteins fold into the free energy minimum structures, but Go further considered that local and non-local interactions in the free energy minimum structures are consistent with each other. Guided by the principle, we proposed a set of rules for designing ideal protein structures stabilized by consistent local and non-local interactions. The rules made possible designs of amino acid sequences with funnel-shaped energy landscapes toward our desired target structures. So far, various protein structures have been created using the rules, which demonstrates significance of our rules as intended. In this review, we briefly describe how the consistency principle impacts on our efforts for developing the design technology.
- 著者
- Yasumasa Joti Akio Kitao
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.16, pp.240-247, 2019 (Released:2019-11-29)
- 参考文献数
- 28
- 被引用文献数
- 2
Terahertz time-domain spectra (THz-TDS) were investigated using the results of molecular dynamics (MD) simulations of Staphylococcal nuclease at two hydration states in the temperature range between 100 and 300 K. The temperature dependence of THz-TDS was found to differ significantly from that of the incoherent neutron scattering spectra (INSS) calculated from the same MD simulation results. We further examined contributions of the mutual and auto-correlations of the atomic fluctuations to THz-TDS and found that the negative value of the former contribution nearly canceled out the positive value of the latter, resulting in a monotonic increase of the reduced absorption cross section. Because of this cancellation, no distinct broad peak was observed in the absorption lineshape function of THz-TDS, whereas the protein boson peak was observed in INSS. The contribution of water molecules to THz-TDS was extremely large for the hydrated protein at temperatures above 200 K, in which large-amplitude motions of water were excited. The combination of THz-TDS, INSS and MD simulations has the potential to extract function-relevant protein dynamics occurring on the picosecond to nanosecond timescale.
- 著者
- Kazuhiro Takemura Akio Kitao
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.16, pp.295-303, 2019 (Released:2019-11-29)
- 参考文献数
- 36
- 被引用文献数
- 4
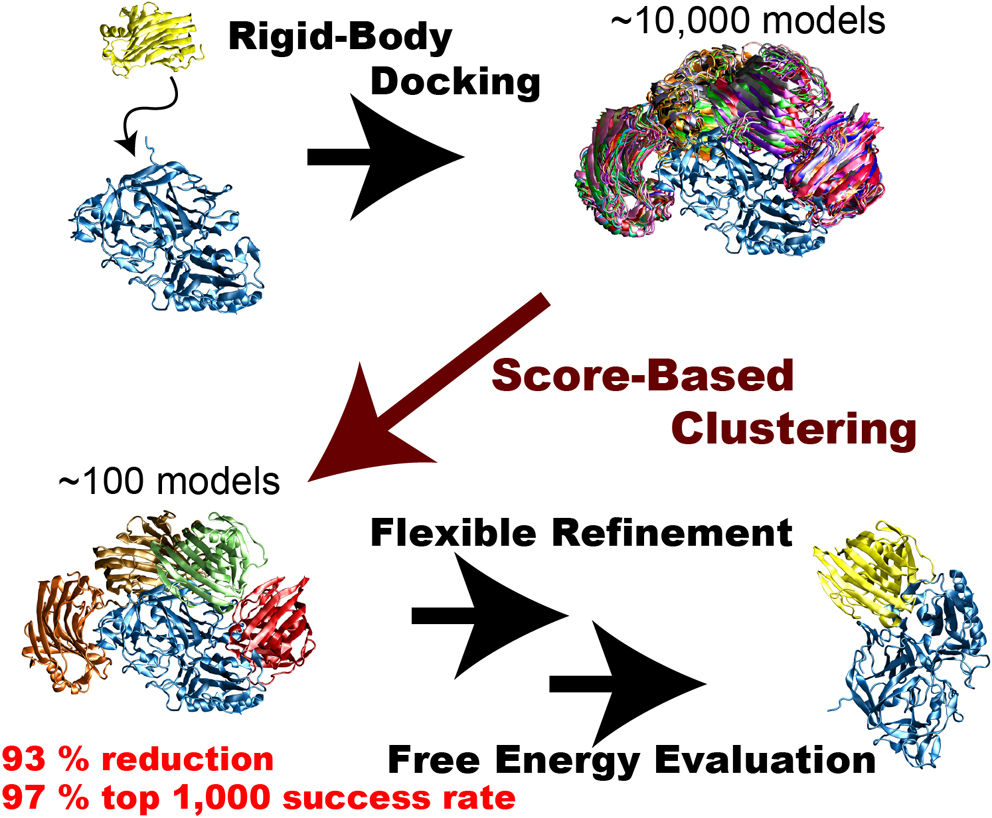
Rigid-body protein-protein docking is very efficient in generating tens of thousands of docked complex models (decoys) in a very short time without considering structure change upon binding, but typical docking scoring functions are not necessarily sufficiently accurate to narrow these decoys down to a small number of plausible candidates. Flexible refinements and sophisticated evaluation of the decoys are thus required to achieve more accurate prediction. Since this process is time-consuming, an efficient screening method to reduce the number of decoys is necessary immediately following rigid-body dockings. We attempted to develop an efficient screening method by clustering decoys generated by the rigid-body docking ZDOCK. We introduced the three metrics ligand-root-mean-square deviation (L-RMSD), interface-ligand-RMSD (iL-RMSD), and the fraction of common contacts (FCC), and examined various ranges of cut-offs for clusters to determine the best set of clustering parameters. Although the employed clustering algorithm is simple, it successfully reduced the number of decoys. Using iL-RMSD with a cut-off radius of 8 Å, the number of decoys that contain at least one near-native model with 90% probability decreased from 4,808 to 320, a 93% reduction in the original number of decoys. Using FCC for the clustering step, the top 1,000 success rates, defined as the probability that the top 1,000 models contain at least one near-native structure, reached 97%. We conclude that the proposed method is very efficient in selecting a small number of decoys that include near-native decoys.
- 著者
- Hiroyuki Terashima Akihiro Kawamoto Yusuke V. Morimoto Katsumi Imada Tohru Minamino
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.14, pp.191-198, 2017 (Released:2017-12-19)
- 参考文献数
- 57
- 被引用文献数
- 1 43
The bacterial flagellum is a supramolecular motility machine consisting of the basal body as a rotary motor, the hook as a universal joint, and the filament as a helical propeller. Intact structures of the bacterial flagella have been observed for different bacterial species by electron cryotomography and subtomogram averaging. The core structures of the basal body consisting of the C ring, the MS ring, the rod and the protein export apparatus, and their organization are well conserved, but novel and divergent structures have also been visualized to surround the conserved structure of the basal body. This suggests that the flagellar motors have adapted to function in various environments where bacteria live and survive. In this review, we will summarize our current findings on the divergent structures of the bacterial flagellar motor.
1 0 0 0 OA Bottom-up creation of cell-free molecular systems: Basic research toward social implementation
- 著者
- Izuru Kawamura Ryuji Kawano Tomoaki Matsuura
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.e200042, (Released:2023-11-10)
1 0 0 0 OA SATORI: Amplification-free digital RNA detection method for the diagnosis of viral infections
- 著者
- Tatsuya Iida Hajime Shinoda Rikiya Watanabe
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.20, no.3, pp.e200031, 2023 (Released:2023-07-22)
- 参考文献数
- 21
- 被引用文献数
- 2
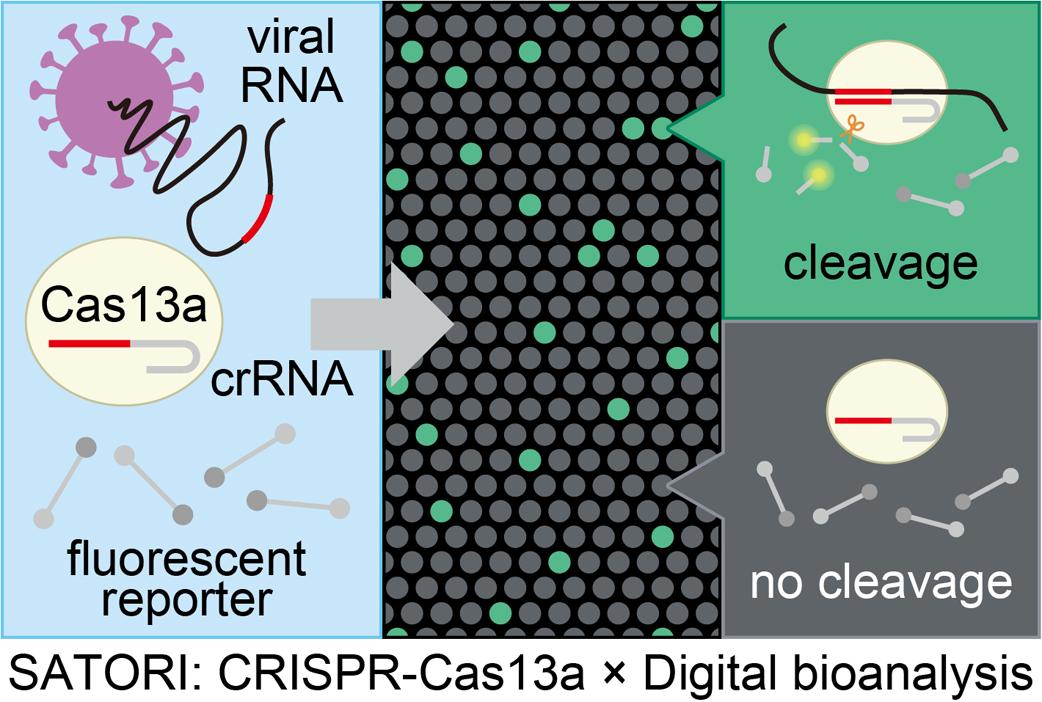
With the recent global outbreak of COVID-19, there is an urgent need to establish a versatile diagnostic method for viral infections. Gene amplification test or antigen test are widely used to diagnose viral infections; however, these methods generally have technical drawbacks either in terms of sensitivity, accuracy, or throughput. To address this issue, we recently developed an amplification-free digital RNA detection method (SATORI), which can identify and detect viral genes at the single-molecule level in approximately 9 min, satisfying almost all detection performance requirements for the diagnosis of viral infections. In addition, we also developed practical platforms for SATORI, such as an automated platform (opn-SATORI) and a low-cost compact fluorescence imaging system (COWFISH), with the aim of application in clinical settings. Our latest technologies can be inherently applied to diagnose a variety of RNA viral infections, such as COVID-19 and Influenza A/B, and therefore, we expect that SATORI will be established as a versatile platform for point-of-care testing of a wide range of infectious diseases, thus contributing to the prevention of future epidemics. This article is an extended version of the Japanese article published in the SEIBUTSU BUTSURI Vol. 63, p. 115–118 (2023).
1 0 0 0 OA Rocking Out Biophysics in IUPAB2024 Kyoto!
- 著者
- Hiroyuki Noji
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.e200039, (Released:2023-10-17)
1 0 0 0 OA The third Japan-U.S. symposium on motor proteins and associated single-molecule biophysics
- 著者
- Tomohiro Shima Kumiko Hayashi
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.e200037, (Released:2023-10-04)
- 著者
- Mika Sakamoto Hirofumi Suzuki Kei Yura
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.16, pp.68-79, 2019 (Released:2019-02-15)
- 参考文献数
- 41
- 被引用文献数
- 3 4
Transport of small molecules across the cell membrane is a crucial biological mechanism for the maintenance of the cell activity. ABC transporter family is a huge group in the transporter membrane proteins and actively transports the substrates using the energy derived from ATP hydrolysis. In humans, there are 48 distinct genes for ABC transporters. A variation of a single amino acid in the amino acid sequence of ABC transporter has been known to be linked with certain disease. The mechanism of the onset of the disease by the variation is, however, still unclear. Recent progress in the method to measure the structures of huge membrane proteins has enabled determination of the 3D structures of ABC transporters and the accumulation of coordinate data of ABC transporter has enabled us to obtain clues for the onset of the disease caused by a single variation of amino acid residue. We compared the structures of ABC transporter in apo and ATP-binding forms and found a possible conformation shift around pivot-like residues in the transmembrane domains. When this conformation change in ABC transporter and the location of pathogenic variation were compared, we found a reasonable match between the two, explaining the onset of the disease by the variation. They likely cause impairment of the pivot-like movement, weakening of ATP binding and weakening of membrane surface interactions. These findings will give a new interpretation of the variations on ABC transporter genes and pave a way to analyse the effect of variation on protein structure and function.
- 著者
- Damien Simon Atsushi Mukaiyama Yoshihiko Furuike Shuji Akiyama
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.19, pp.e190008, 2022 (Released:2022-04-14)
- 参考文献数
- 49
- 被引用文献数
- 1 5
KaiC is the central pacemaker of the circadian clock system in cyanobacteria and forms the core in the hetero-multimeric complexes, such as KaiB–KaiC and KaiA–KaiB–KaiC. Although the formation process and structure of the binary and ternary complexes have been studied extensively, their disassembly dynamics have remained elusive. In this study, we constructed an experimental system to directly measure the autonomous disassembly of the KaiB–KaiC complex under the condition where the dissociated KaiB cannot reassociate with KaiC. At 30°C, the dephosphorylated KaiB–KaiC complex disassembled with an apparent rate of 2.1±0.3 d–1, which was approximately twice the circadian frequency. Our present analysis using a series of KaiC mutants revealed that the apparent disassembly rate correlates with the frequency of the KaiC phosphorylation cycle in the presence of KaiA and KaiB and is robustly temperature-compensated with a Q10 value of 1.05±0.20. The autonomous cancellation of the interactions stabilizing the KaiB–KaiC interface is one of the important phenomena that provide a link between the molecular-scale and system-scale properties.
- 著者
- Kazusa Beppu Yusuke T. Maeda
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.19, pp.e190020, 2022 (Released:2022-06-03)
- 参考文献数
- 25
- 被引用文献数
- 1
Ordered collective motion emerges in a group of autonomously motile elements (known as active matter) as their density increases. Microswimmers, such as swimming bacteria, have been extensively studied in physics and biology. A dense suspension of bacteria forms seemingly chaotic turbulence in viscous fluids. Interestingly, this active turbulence driven by bacteria can form a hidden ensemble of many vortices. Understanding the active turbulence in a bacterial suspension can provide physical principles for pattern formation and insight into the instability underlying biological phenomena. This review presents recent findings regarding ordered structures causing active turbulence and discusses a physical approach for controlling active turbulence via geometric confinement. When the active matter is confined in a compartment with a size comparable to the correlation length of the collective motion, vortex-like rotation appears, and the vortex pairing order is indicated by the patterns of interacting vortices. Additionally, we outline the design principle for controlling collective motions via the geometric rule of the vortex pairing, which may advance engineering microdevices driven by a group of active matter. This article is an extended version of the Japanese article, Ordered Structure and Geometric Control of Active Matter in Dense Bacterial Suspensions, published in SEIBUTSU BUTSURI Vol. 60, p. 13–18 (2020).
- 著者
- Tohru Minamino Miki Kinoshita Yusuke V. Morimoto Keiichi Namba
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.19, pp.e190046, 2022 (Released:2022-12-07)
- 参考文献数
- 83
- 被引用文献数
- 3
Bacteria employ the flagellar type III secretion system (fT3SS) to construct flagellum, which acts as a supramolecular motility machine. The fT3SS of Salmonella enterica serovar Typhimurium is composed of a transmembrane export gate complex and a cytoplasmic ATPase ring complex. The transmembrane export gate complex is fueled by proton motive force across the cytoplasmic membrane and is divided into four distinct functional parts: a dual-fuel export engine; a polypeptide channel; a membrane voltage sensor; and a docking platform. ATP hydrolysis by the cytoplasmic ATPase complex converts the export gate complex into a highly efficient proton (H+)/protein antiporter that couples inward-directed H+ flow with outward-directed protein export. When the ATPase ring complex does not work well in a given environment, the export gate complex will remain inactive. However, when the electric potential difference, which is defined as membrane voltage, rises above a certain threshold value, the export gate complex becomes an active H+/protein antiporter to a considerable degree, suggesting that the export gate complex has a voltage-gated activation mechanism. Furthermore, the export gate complex also has a sodium ion (Na+) channel to couple Na+ influx with flagellar protein export. In this article, we review our current understanding of the activation mechanism of the dual-fuel protein export engine of the fT3SS. This review article is an extended version of a Japanese article, Membrane voltage-dependent activation of the transmembrane export gate complex in the bacterial flagellar type III secretion system, published in SEIBUTSU BUTSURI Vol. 62, p165–169 (2022).