- 著者
- Tony Z. Jia Yutetsu Kuruma
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.bppb-v18.032, (Released:2021-11-18)
- 著者
- Izuru Kawamura Hayato Seki Seiya Tajima Yoshiteru Makino Arisu Shigeta Takashi Okitsu Akimori Wada Akira Naito Yuki Sudo
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.bppb-v18.019, (Released:2021-07-14)
- 被引用文献数
- 6
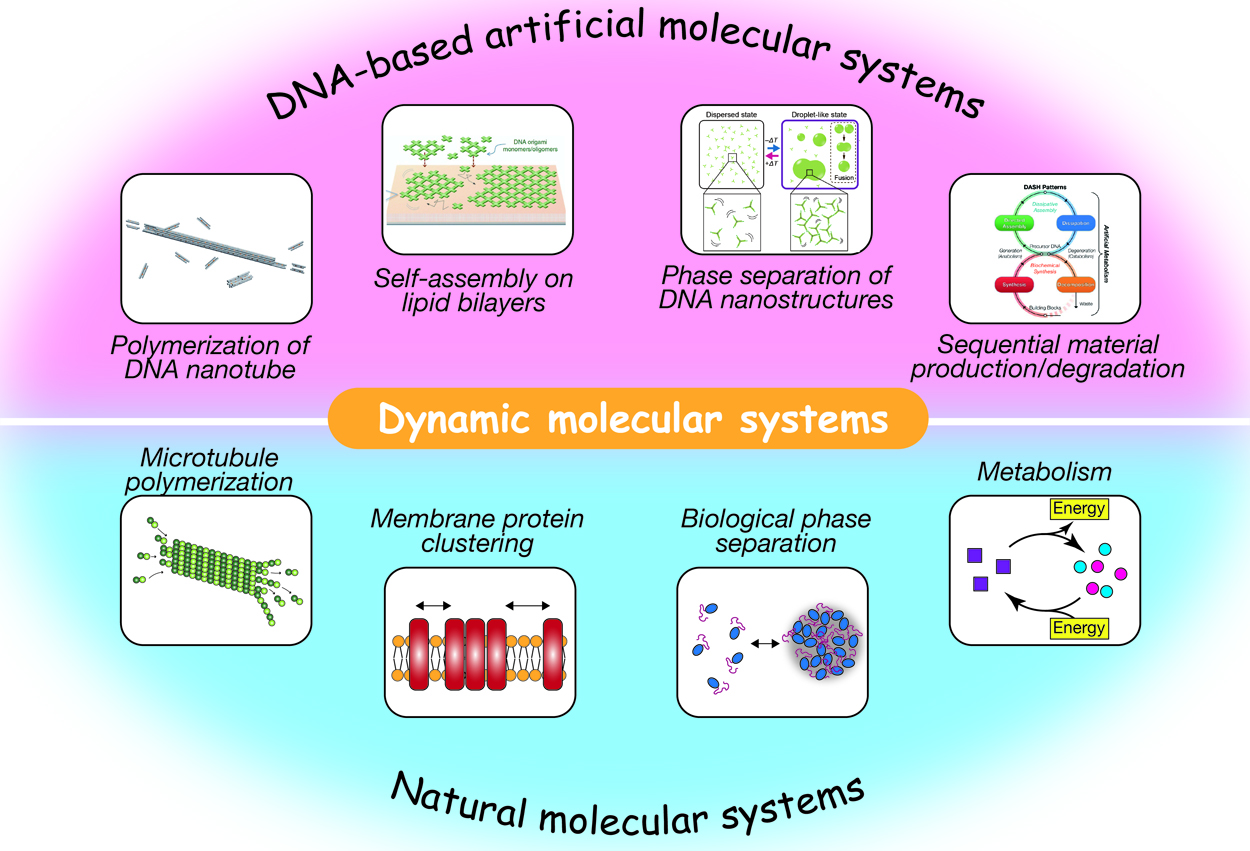
3 0 0 0 OA DNA nanotechnology provides an avenue for the construction of programmable dynamic molecular systems
- 著者
- Yusuke Sato Yuki Suzuki
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.18, pp.116-126, 2021 (Released:2021-05-26)
- 参考文献数
- 80
- 被引用文献数
- 2
Self-assembled supramolecular structures in living cells and their dynamics underlie various cellular events, such as endocytosis, cell migration, intracellular transport, cell metabolism, and gene expression. Spatiotemporally regulated association/dissociation and generation/degradation of assembly components is one of the remarkable features of biological systems. The significant advancement in DNA nanotechnology over the last few decades has enabled the construction of various-shaped nanostructures via programmed self-assembly of sequence-designed oligonucleotides. These nanostructures can further be assembled into micrometer-sized structures, including ordered lattices, tubular structures, macromolecular droplets, and hydrogels. In addition to being a structural material, DNA is adopted to construct artificial molecular circuits capable of activating/inactivating or producing/decomposing target DNA molecules based on strand displacement or enzymatic reactions. In this review, we provide an overview of recent studies on artificially designed DNA-based self-assembled systems that exhibit dynamic features, such as association/dissociation of components, phase separation, stimulus responsivity, and DNA circuit-regulated structural formation. These biomacromolecule-based, bottom-up approaches for the construction of artificial molecular systems will not only throw light on bio-inspired nano/micro engineering, but also enable us to gain insights into how autonomy and adaptability of living systems can be realized.
- 著者
- Akihiko Nakamura Kei-ichi Okazaki Tadaomi Furuta Minoru Sakurai Jun Ando Ryota Iino
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.BSJ-2020004, (Released:2020-06-09)
- 被引用文献数
- 5
- 著者
- Jean-François Gibrat
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.16, pp.444-451, 2019 (Released:2019-11-29)
- 参考文献数
- 13
This paper presents a preliminary work consisting of two contributions. The first one is the design of a very efficient algorithm based on an “Overlap-Layout-Consensus” (OLC) graph to assemble the long reads provided by 3rd generation technologies. The second concerns the analysis of this graph using algebraic topology concepts to determine, in advance, whether the assembly of the genome will be straightforward, i.e., whether it will lead to a pseudo-Hamiltonian path or cycle, or whether the results will need to be scrutinized. In the latter case, it will be necessary to look for “loops” in the OLC assembly graph caused by unresolved repeated genomic regions, and then try to untie the “knots” created by these regions.
- 著者
- Junko Taguchi Akio Kitao
- 出版者
- 一般社団法人 日本生物物理学会
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.13, pp.117-126, 2016 (Released:2016-07-14)
- 参考文献数
- 37
- 被引用文献数
- 7
We examine the dynamic features of non-trivial allosteric binding sites to elucidate potential drug binding sites. These allosteric sites were previously found to be allosteric after determination of the protein-drug co-crystal structure. After comprehensive search in the Protein Data Bank, we identify 10 complex structures with allosteric ligands whose structures are very similar to their functional forms. Then, possible pockets on the protein surface are searched as potential ligand binding sites. To mimic ligand binding to the pocket, complex models are generated to fill out each pocket with pseudo ligand blocks consisting of spheres. Normal mode analysis of the elastic network model is performed for the complex models and unbound structures to assess the change of protein dynamics induced by ligand binding. We examine nine profiles to describe the dynamic and positional characteristics of the pockets, and identify the change of fluctuation around the ligand, ΔMSFbs, as the best profile for distinguishing the allosteric sites from the other sites in 8 structures. These cases should be considered as examples of dynamics-driven allostery, which accompanies significant changes in protein dynamics. ΔMSFbs is suggested to be used for the search of potential dynamics-driven allosteric sites in proteins for drug discovery.
- 著者
- Akihiro Okamoto Yoshihide Tokunou Junki Saito
- 出版者
- 一般社団法人 日本生物物理学会
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.13, pp.71-76, 2016 (Released:2016-05-27)
- 参考文献数
- 32
- 被引用文献数
- 11
Outer-membrane c-type cytochrome (OM c-Cyt) complexes in several genera of iron-reducing bacteria, such as Shewanella and Geobacter, are capable of transporting electrons from the cell interior to extracellular solids as a terminal step of anaerobic respiration. The kinetics of this electron transport has implications for controlling the rate of microbial electron transport during bioenergy or biochemical production, iron corrosion, and natural mineral cycling. Herein, we review the findings from in-vivo and in-vitro studies examining electron transport kinetics through single OM c-Cyt complexes in Shewanella oneidensis MR-1. In-vitro electron flux via a purified OM c-Cyt complex, comprised of MtrA, B, and C proteins from S. oneidensis MR-1, embedded in a proteoliposome system is reported to be 10- to 100-fold faster compared with in-vivo estimates based on measurements of electron flux per cell and OM c-Cyts density. As the proteoliposome system is estimated to have 10-fold higher cation flux via potassium channels than electrons, we speculate that the slower rate of electron-coupled cation transport across the OM is responsible for the significantly lower electron transport rate that is observed in-vivo. As most studies to date have primarily focused on the energetics or kinetics of interheme electron hopping in OM c-Cyts in this microbial electron transport mechanism, the proposed model involving cation transport provides new insight into the rate detemining step of EET, as well as the role of self-secreted flavin molecules bound to OM c-Cyt and proton management for energy conservation and production in S. oneidensis MR-1.
- 著者
- Ryota Sakamoto Yusuke T. Maeda
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.20, no.3, pp.e200032, 2023 (Released:2023-09-01)
- 参考文献数
- 52
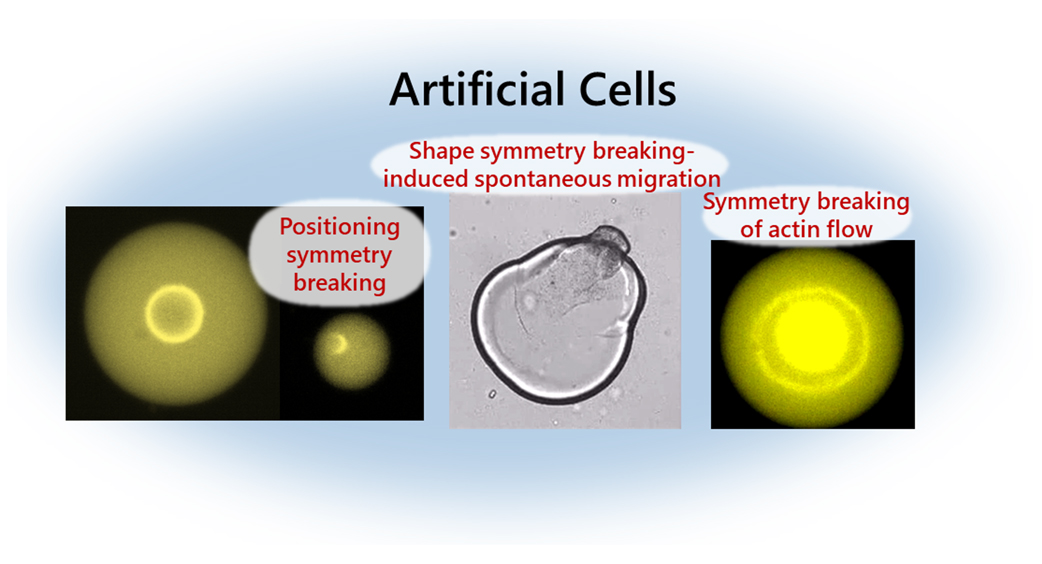
Single-cell behaviors cover many biological functions, such as cell division during morphogenesis and tissue metastasis, and cell migration during cancer cell invasion and immune cell responses. Symmetry breaking of the positioning of organelles and the cell shape are often associated with these biological functions. One of the main players in symmetry breaking at the cellular scale is the actin cytoskeleton, comprising actin filaments and myosin motors that generate contractile forces. However, because the self-organization of the actomyosin network is regulated by the biochemical signaling in cells, how the mechanical contraction of the actin cytoskeleton induces diverse self-organized behaviors and drives the cell-scale symmetry breaking remains unclear. In recent times, to understand the physical underpinnings of the symmetry breaking exhibited in the actin cytoskeleton, artificial cell models encapsulating the cytoplasmic actomyosin networks covered with lipid monolayers have been developed. By decoupling the actomyosin mechanics from the complex biochemical signaling within living cells, this system allows one to study the self-organization of actomyosin networks confined in cell-sized spaces. We review the recent developments in the physics of confined actomyosin networks and provide future perspectives on the artificial cell-based approach. This review article is an extended version of the Japanese article, The Physical Principle of Cell Migration Under Confinement: Artificial Cell-based Bottom-up Approach, published in SEIBUTSU BUTSURI Vol. 63, p. 163–164 (2023).
2 0 0 0 OA Announcement of BPPB paper awards 2023
- 著者
- Haruki Nakamura
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.20, no.4, pp.e200040, 2023 (Released:2023-11-07)
- 参考文献数
- 8
2 0 0 0 OA Frontiers of microbial movement research
- 著者
- Tohru Minamino Daisuke Nakane Shuichi Nakamura Hana Kiyama Yusuke V. Morimoto Makoto Miyata
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.20, no.3, pp.e200033, 2023 (Released:2023-09-15)
- 参考文献数
- 19
- 著者
- Hideaki Yoshimura
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.19, pp.e190007, 2022 (Released:2022-03-26)
- 参考文献数
- 43
Membrane receptors provide interfaces of various extracellular stimuli to transduce the signal into the cell. Receptors are required to possess such conflicting properties as high sensitivity and noise reduction for the cell to keep its homeostasis and appropriate responses. To understand the mechanisms by which these functions are achieved, single-molecule monitoring of the motilities of receptors and signaling molecules on the plasma membrane is one of the most direct approaches. This review article introduces several recent single-molecule imaging studies of receptors, including the author’s recent work on triple-color single-molecule imaging of G protein-coupled receptors. Based on these researches, advantages and perspectives of the single-molecule imaging approach to solving the mechanisms of receptor functions are illustrated.
2 0 0 0 OA Removing the parachuting artifact using two-way scanning data in high-speed atomic force microscopy
- 著者
- Shintaroh Kubo Kenichi Umeda Noriyuki Kodera Shoji Takada
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.20, no.1, pp.e200006, 2023 (Released:2023-02-14)
- 参考文献数
- 29
- 被引用文献数
- 4
The high-speed atomic force microscopy (HS-AFM) is a unique and prominent method to observe structural dynamics of biomolecules at single molecule level at near-physiological condition. To achieve high temporal resolution, the probe tip scans the stage at high speed which can cause the so-called parachuting artifact in the HS-AFM images. Here, we develop a computational method to detect and remove the parachuting artifact in HS-AFM images using the two-way scanning data. To merge the two-way scanning images, we employed a method to infer the piezo hysteresis effect and to align the forward- and backward-scanning images. We then tested our method for HS-AFM videos of actin filaments, molecular chaperone, and duplex DNA. Together, our method can remove the parachuting artifact from the raw HS-AFM video containing two-way scanning data and make the processed video free from the parachuting artifact. The method is general and fast so that it can easily be applied to any HS-AFM videos with two-way scanning data.
- 著者
- Kentaro Ozawa Hirotaka Taomori Masayuki Hoshida Ituki Kunita Sigeru Sakurazawa Hajime Honda
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.16, pp.1-8, 2019 (Released:2019-01-09)
- 参考文献数
- 27
- 被引用文献数
- 2 4
The movements of single actin filaments along a myosin-fixed glass surface were observed under a conventional fluorescence microscope. Although random at a low concentration, moving directions of filaments were aligned by the presence of over 1.0 mg/mL of unlabeled filaments. We found that actin filaments when at the intermediate concentrations ranging from 0.1 to 1.0 mg/mL, formed winding belt-like patterns and moved in a two-directional manner along the belts. These patterns were spread over a millimeter range and found to have bulged on the glass in a three-dimensional manner. Filaments did not get closer than about 37.5 nm to each other within each belt-pattern. The average width and the curvature radius of the pattern did not apparently change even when the range of actin concentrations was between 0.05 and 1.0 mg/mL or the sliding velocity between 1.2 and 3.2 μm/sec. However, when the length of filaments was shortened by ultrasonic treatments or the addition of gelsolin molecules, the curvature radius became small from 100 to 60 μm. These results indicate that this belt-forming nature of actin filaments may be due to some inter-filament interactions.
- 著者
- Seine A. Shintani
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.18, pp.85-95, 2021 (Released:2021-04-16)
- 参考文献数
- 38
- 被引用文献数
- 5
The effects of high pressure (40–70 MPa) on the structure and function of myofibrils were investigated by high pressure microscopy. When this pressure was applied to myofibrils immersed in relaxing solution, the sarcomere length remained almost unchanged, and the A band became shorter and wider. The higher the applied pressure, the faster the change. However, shortening and widening of the A band were not observed when pressure was applied to myofibrils immersed in a solution obtained by omitting ATP from the relaxing solution. However, even under these conditions, structural loss, such as loss of the Z-line structure, occurred. In order to evaluate the consequences of this pressure-treated myofibril, the oscillatory movement of sarcomere (sarcomeric oscillation) was evoked and observed. It was possible to induce sarcomeric oscillation even in pressure-treated myofibrils whose structure was destroyed. The pressurization reduced the total power of the sarcomeric oscillation, but did not change the average frequency. The average frequency did not change even when a pressure of about 40 MPa was applied during sarcomeric oscillation. The average frequency returned to the original when the pressure was returned to the original value after applying stronger pressure to prevent the sarcomere oscillation from being observed. This result suggests that the decrease in the number of myosin molecules forming the crossbridge does not affect the average frequency of sarcomeric oscillation. This fact will help build a mechanical hypothesis for sarcomeric oscillation. The pressurization treatment is a unique method for controlling the structure of myofibrils as described above.
- 著者
- Takuya Yoshizawa Hiroyoshi Matsumura
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.17, pp.25-29, 2020-04-04 (Released:2020-04-04)
- 参考文献数
- 48
- 被引用文献数
- 2 5
Low-complexity (LC) sequences, regions that are predominantly made up of limited amino acids, are often observed in eukaryotic nuclear proteins. The role of these LC sequences has remained unclear for decades. Recent studies have shown that LC sequences are important in the formation of membrane-less organelles via liquid–liquid phase separation (LLPS). The RNA binding protein, fused in sarcoma (FUS), is the most widely studied of the proteins that undergo LLPS. It forms droplets, fibers, or hydrogels using its LC sequences. The N-terminal LC sequence of FUS is made up of Ser, Tyr, Gly, and Gln, which form a labile cross-β polymer core while the C-terminal Arg-Gly-Gly repeats accelerate LLPS. Normally, FUS localizes to the nucleus via the nuclear import receptor karyopherin β2 (Kapβ2) with the help of its C-terminal proline-tyrosine nuclear localization signal (PY-NLS). Recent findings revealed that Kapβ2 blocks FUS mediated LLPS, suggesting that Kapβ2 is not only a transport protein but also a chaperone which regulates LLPS during the formation of membrane-less organelles. In this review, we discuss the effects of the nuclear import receptors on LLPS.
2 0 0 0 OA Mathematical model of chromosomal dynamics during DNA double strand break repair in budding yeast
- 著者
- Shinjiro Nakahata Tetsushi Komoto Masashi Fujii Akinori Awazu
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.19, pp.e190012, 2022 (Released:2022-04-20)
- 参考文献数
- 40
- 被引用文献数
- 1
During the repair of double-strand breaks (DSBs) in DNA, active mobilizations for conformational changes in chromosomes have been widely observed in eukaryotes, from yeast to animal and plant cells. DSB-damaged loci in the yeast genome showed increased mobility and relocation to the nuclear periphery. However, the driving forces behind DSB-induced chromatin dynamics remain unclear. In this study, mathematical models of normal and DSB-damaged yeast chromosomes were developed to simulate their structural dynamics. The effects of histone degradation in the whole nucleus and the change in the physical properties of damaged loci due to the binding of SUMOylated repair proteins were considered in the model of DSB-induced chromosomes based on recent experimental results. The simulation results reproduced DSB-induced changes to structural and dynamical features by which the combination of whole nuclear histone degradation and the rigid structure formation of repair protein accumulations on damaged loci were suggested to be primary contributors to the process by which damaged loci are relocated to the nuclear periphery.
2 0 0 0 OA Molecular mechanisms of amyloid-β peptide fibril and oligomer formation: NMR-based challenges
- 著者
- Hidekazu Hiroaki
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.20, no.1, pp.e200007, 2023 (Released:2023-03-04)
- 参考文献数
- 42
- 被引用文献数
- 1
To completely treat and ultimately prevent dementia, it is essential to elucidate its pathogenic mechanisms in detail. There are two major hypotheses for the pathogenesis of Alzheimer’s dementia: the β-amyloid (Aβ) hypothesis and the tau hypothesis. The modified amyloid hypothesis, which proposes that toxic oligomers rather than amyloid fibrils are the essential cause, has recently emerged. Aβ peptides [Aβ(1–40) and Aβ(1–42)] form highly insoluble aggregates in vivo and in vitro. These Aβ aggregates contain many polymorphisms, whereas Aβ peptides are intrinsically disordered in physiological aqueous solutions without any compact conformers. Over the last three decades, solid-state nuclear magnetic resonance (NMR) has greatly contributed to elucidating the structure of each polymorph, while solution NMR has revealed the dynamic nature of the transient conformations of the monomer. Moreover, several methods to investigate the aggregation process based on the observation of magnetization saturation transfer have also been developed. The complementary use of NMR methods with cryo-electron microscopy, which has rapidly matured, is expected to clarify the relationship between the amyloid and molecular pathology of Alzheimer’s dementia in the near future. This review article is an extended version of the Japanese article, Insights into the Mechanisms of Oligomerization/Fibrilization of Amyloid β Peptide from Nuclear Magnetic Resonance, published in SEIBUTSU BUTSURI Vol. 62, p. 39–42 (2022).
- 著者
- Etsuro Ito Rei Shima Tohru Yoshioka
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.16, pp.132-139, 2019 (Released:2019-08-24)
- 参考文献数
- 79
- 被引用文献数
- 26
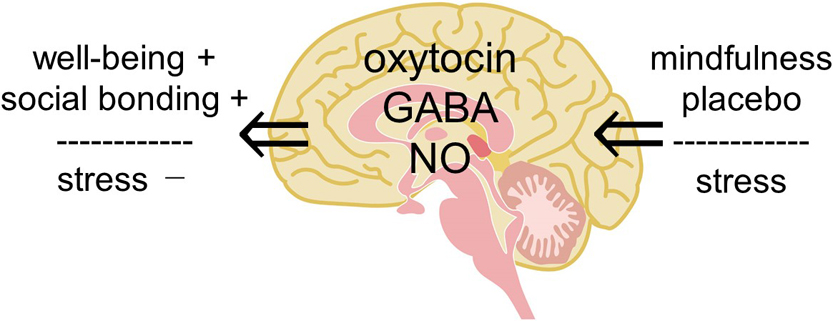
We review the involvement of a small molecule, oxytocin, in various effects of physical stimulation of somatosensory organs, mindfulness meditation, emotion and fragrance on humans, and then propose a hypothesis that complex human states and behaviors, such as well-being, social bonding, and emotional behavior, are explained by oxytocin. We previously reported that oxytocin can induce pain relief and described the possibility how oxytocin in the dorsal horn and/or the dorsal root ganglion relieves joint and muscle pain. In the present article, we expand our research target from the physical analgesic effects of oxytocin to its psychologic effects to upregulate well-being and downregulate stress and anxiety. For this purpose, we propose a “hypothalamic-pituitary-adrenal (HPA) axis-oxytocin model” to explain why mindfulness meditation, placebo, and fragrance can reduce stress and anxiety, resulting in contentment. This new proposed model of HPA axis-oxytocin in the brain also provides a target to address other questions regarding emotional behaviors, learning and memory, and excess food intake leading to obesity, aimed at promoting a healthy life.
- 著者
- Kenta Odagiri Hiroshi Fujisaki Hiroya Takada Rei Ogawa
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- pp.e200023, (Released:2023-05-24)
- 被引用文献数
- 2
To computationally investigate the recent experimental finding such that extracellular ATP release caused by exogeneous mechanical forces promote wound closure, we introduce a mathematical model, the Cellular Potts Model (CPM), which is a popular discretized model on a lattice, where the movement of a “cell” is determined by a Monte Carlo procedure. In the experiment, it was observed that there is mechanosensitive ATP release from the leading cells facing the wound gap and the subsequent extracellular Ca2+ influx. To model these phenomena, the Reaction-Diffusion equations for extracellular ATP and intracellular Ca2+ concentrations are adopted and combined with CPM, where we also add a polarity term because the cell migration is enhanced in the case of ATP release. From the numerical simulations using this hybrid model, we discuss effects of the collective cell migration due to the ATP release and the Ca2+ influx caused by the mechanical forces and the consequent promotion of wound closure.
- 著者
- Jonathan R. Church Jógvan Magnus Haugaard Olsen Igor Schapiro
- 出版者
- The Biophysical Society of Japan
- 雑誌
- Biophysics and Physicobiology (ISSN:21894779)
- 巻号頁・発行日
- vol.20, no.Supplemental, pp.e201007, 2023 (Released:2023-03-21)
- 参考文献数
- 50
- 被引用文献数
- 1
Multiscale simulations have been established as a powerful tool to calculate and predict excitation energies in complex systems such as photoreceptor proteins. In these simulations the chromophore is typically treated using quantum mechanical (QM) methods while the protein and surrounding environment are described by a classical molecular mechanics (MM) force field. The electrostatic interactions between these regions are often treated using electrostatic embedding where the point charges in the MM region polarize the QM region. A more sophisticated treatment accounts also for the polarization of the MM region. In this work, the effect of such a polarizable embedding on excitation energies was benchmarked and compared to electrostatic embedding. This was done for two different proteins, the lipid membrane-embedded jumping spider rhodopsin and the soluble cyanobacteriochrome Slr1393g3. It was found that the polarizable embedding scheme produces absorption maxima closer to experimental values. The polarizable embedding scheme was also benchmarked against expanded QM regions and found to be in qualitative agreement. Treating individual residues as polarizable recovered between 50% and 71% of the QM improvement in the excitation energies, depending on the system. A detailed analysis of each amino acid residue in the chromophore binding pocket revealed that aromatic residues result in the largest change in excitation energy compared to the electrostatic embedding. Furthermore, the computational efficiency of polarizable embedding allowed it to go beyond the binding pocket and describe a larger portion of the environment, further improving the results.